Antibiotic resistance fate in the full-scale drinking water and municipal wastewater treatment processes: A review
Article information
Abstract
Antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) have been widely identified as emerging pollutants in various aquatic compartments. Concerns have been raised that the antibiotic resistance determinants may occur in treated drinking water and wastewater, weakening the therapeutic efficacy of antibiotics and so posing threat to public health. Most often, laboratory trials are conducted to assess the removal efficiency of ARB and ARGs in individual treatment processes (e.g., disinfection). However, the random variations of microbial distribution and chemical compositions in real-world environments cannot be entirely simulated, presumably leading to false-positive results as a consequence. It is therefore useful to provide a summary of recent advancements regarding the antibiotic resistance attenuation during full-scale water/wastewater treatments, which has not been adequately evaluated so far. In this review, the prevalence, proliferation and transmission of ARB and ARGs in urban water cycle, coupled with corresponding detection methods are presented as a short overview. The fate of ARB and ARGs in the sequential drinking water and wastewater processing units is critically summarized, the aim of which is to provide guidance for improving the current water treatment facilities to further reduce the antibiotic resistance in finished water.
1. Introduction
Antibiotics are extensively used as the first choice to combat bacterial infections in humans and animals. Recent modelling studies suggested that the global consumption of antibiotics would increase 161% between 2015 and 2030, from 42.3 billion to 68.1 billion, expressed in defined daily doses [1]. After being incomplete metabolized, about half of the consumed antibiotics are excreted and enter the urban water cycle [2]. Due to the incomplete removal by urban wastewater treatment plants (UWTPs), the remaining chemicals can be constantly accumulated in natural aquatic environment [3]. As reviewed by Liu et al. [4], the maximum concentrations of 37 common antibiotics in typical China lakes reached 940 ng/L. Consequently, the indigenous microbial communities exposed to long-term selective pressure gradually acquire resistance or activate their intrinsic resistance to specific antibiotics [5, 6], and this poses an upsurge of investigations on the antibiotic resistance determinants. Incorporated by antibiotic resistant bacteria (ARB), the antibiotic resistance genes (ARGs) can be widely transferred from donors to recipients by gene transfer (parental or non-parental), and can be life-threatening if integrated by the pathogenic bacteria accessible to human beings [7]. For instance, methicillin-resistant Staphylococcus aureus (MRSA) is always a main culprit of infection in the community settings and the Center for Disease Dynamics, and Economics & Policy (CDDEP) in US reported increasing numbers of MRSA hospitalizations in recent years [8]. World Health Organization (WHO) also proclaimed that antibiotic resistance is growing, and we are fast running out of treatment options for bacterial infections [9].
Aqueous media are ideal for the proliferation and transmission of antibiotic resistance determinants [10]. Diversified ARB/ARGs have been frequently detected in surface water and groundwater [11, 12], and even in drinking water supplies [13–15]. UWTPs and drinking water treatment plants (DWTPs) are vital barriers in urban water cycle, which typically include sequential processing units to protect both environment and human health [3]. So, it’s very important to know whether these treatment utilities are efficient for antibiotic resistance removal.
Numerous researchers have conducted lab-scale trials to discern the antibiotic resistance attenuation in individual processing units [16, 17], however, they could neither simulate the random variations of autochthonous microbial communities in actual treatment systems, nor elucidate the seasonal and regional distribution of antibiotic resistance [18]. Hence, the results of field trials should be underscored and expanded clarification is still required. On this basis, the present review synthetically evaluates the removal effects of ARB and ARGs in each processing unit, as well as the corresponding mechanisms based on recent investigations in full-scale DWTPs/UWTPs. This paper is divided into four parts: (i) occurrence and transmission of ARB and ARGs in urban water cycle, (ii) common detection methods for ARB and ARGs in aqueous media, (iii) antibiotic resistance fate in full-scale drinking water and (iv) wastewater treatment processes (from raw water to finished water). It is an attempt to provide useful guidance for improving the performance of current resistance interventions.
2. The Prevalence of ARB/ARGs in Urban Water Cycle
The fate of antibiotic resistance in urban water cycle and possible exposures to the humans are depicted in Fig. 1. As anthropogenic pollution sources, hospital wastewater, livestock wastes, community sewage and pharmaceutical sewage contain a multitude of antibiotics as well as ARB/ARGs [19, 20] that are collected or directly discharged. Municipal wastewater with complicated constituents is treated by UWTPs, which decrease the concentrations of antibiotics and resistance determinants to some extent. The remaining antibiotics then enter the effluent-receiving water, where they constantly exert selective pressure on indigenous microorganisms, inducing the resistomes and further promoting ARG transmission [6]. Residual resistance can also migrate into the soil with sludge landfill or diffuse into the air (Fig. 1). Accordingly, the antibiotic resistance determinants are ubiquitous in various aquatic compartments, including drinking water sources [21, 22]. Drinking water treatment is the last resort to reduce the impact of antibiotic resistance on human health, and the remaining ARB/ARGs in final water can be re-enriched in the distribution system and contaminate the tap water [23]. However, antibiotic resistance has not been listed as a target pollutant by water/wastewater treatment facilities [24], and no maximum levels of ARB/ARGs have been set by international institutions to date.

The fate of antibiotics and antibiotic resistance determinants in urban water recycle and their main access to the human beings (thickness of arrow indicates the abundance of antibiotic resistance determinants and the main factors that affect the fate of antibiotic resistance are enclosed by the boxes).
2.1. Transmission Pathways of ARGs in the Water
To survive under the selective pressure caused by active antibiotics, indigenous bacteria tend to incorporate intracellular ARGs (iARGs). There are two pathways through which this may occur: (i) Inheriting ARGs from the parent ARB via vertical gene transfer, and (ii) obtaining ARGs from other accessible sources (such as non-parental ARB, extracellular ARGs (eARGs) or bacteriophages) via horizontal gene transfer (HGT), which enables the acquisition of new genetic materials from outside of clonal lineage and helps the bacteria to rapidly adapt to selective pressure [25], thus promoting ARG transfer between indigenous bacteria and distantly related pathogenic communities. As key vehicles for HGT, mobile genetic elements (MGEs) such as plasmids, integrons, transposons have gained increasing attention due to their ubiquitous and persistent presence in aquatic environments [26]. Studies have shown that phage-borne ARGs could attach to the protective surfaces of colloids and persist for an extended period (up to weeks), during which time further transmission and proliferation would be successfully accomplished [27].
2.2. Antibiotic Resistance Levels in Surface Water
The prevalence of antibiotic resistance determinants, which is closely associated with the types of ARGs and indigenous bacteria communities, has been extensively investigated in surface water. The absolute and relative abundance (normalized by 16S gene copies) of several commonly detected ARGs (sul1, sul2, tetA, tetW, blaTEM, ermB and qnrS) are summarized in Table S1 and Table S2, respectively. The sul1 and sul2 genes encode resistance to sulfonamides, which were introduced in 1937 as the first effective antibiotics to prevent infections in human and animals [28]. As shown in Fig. 2, the mean concentrations of sul1 and sul2 were the highest (105–106 copies/mL) among the seven investigated species in China, which was probably because there were higher accumulation rates of sulfonamides compared to other broad-spectrum antibiotics in the natural surface water [4]. Tetracycline-resistant genes (tetA and tetW) were also ubiquitous in aquatic environments (up to 105 copies/mL). In addition, relatively high levels of blaTEM, ermB and qnrS (up to 104, 105, 108 copies/mL) were also identified, which may have been a result of specific antibiotic consuming patterns.
Geographic variations in the abundance of ARGs in surface water are shown in Fig. 2. The results reveal that sul1, sul2 and ermB are significantly more abundant in China than in other countries (p < 0.05). Moreover, the absolute abundance of sul1 and sul2 in effluent-receiving waters is found to be 2–4 orders of magnitude larger than in low-impact surface water (Table S3), indicating that UWTP discharges somehow increased the antibiotic resistance levels of the receiving water. Hiller et al. [29] reported that the resistance levels were positively correlated with sewage discharge in general, but these findings were contradicted by Schreiber and Kistemann [30], who observed that multidrug resistance of Rhodospirillaceae was attenuated with the increasing sewage discharge. In that case, other pollution sources (such as the direct emission of livestock sewage) might serve as the main contributors of resistance [30]. Interestingly, a lagging increase in the abundance of dominant ARGs and MGEs was identified in the receiving water, even though the effluent of UWTPs was effectively treated and the ARG abundance was lower than that of the receiving water [31]. Therefore, the long-term impact and reasonable emission standards of UWTPs should be further investigated.
2.3. Antibiotic Resistance Levels in the Sources of Drinking Water and Municipal Wastewater
To have a general impression of the target antibiotic resistance levels for DWTPs and UWTPs, the abundance of ARB and ARGs in sources of drinking water and municipal wastewater is summarized as shown in Table 1, Table 2 and Table S4. In the sources of drinking water, bacterial resistance to tetracycline, sulfonamides and ampicillin was commonly identified (Table S4), and sulfonamide-resistant percentages of Escherichia coli (E. coli) isolates reached 100% in some studies [50, 51]. High percentages (35%–70%) of Enterobacteriaceae and E. coli isolates were found to be resistant to norfloxacin. Moreover, studies also highlighted the occurrence of multidrug-resistant isolates. For example, antimicrobial susceptibility tests revealed that 11% of the isolated E. coli from the well water in India were resistant to three or more groups of antibiotics [21].

The Absolute Abundance of Several Common ARGs in the Sources of Drinking Water and Municipal Wastewater (copies/mL)
The absolute abundance of five ARGs (sul1, sul2, tetA, blaTEM and ermB) and the Class I integron integrase gene (intI1) in the sources of drinking water is also summarized in Table 2. Antibiotic resistance integrons are classified into five grades (Class I–Class V) based on the amino acid sequences of the integrase protein encoded by intI. As shown in Table 2, the resistance levels of raw water in full-scale DWTPs were similar to or slightly lower than those in surface water and the absolute abundance of all of the investigated ARGs was lower than 106 copies/mL. The resistance levels of sul genes were comparable in the raw water of DWTPs (from the Yangtze or Huangpu rivers). Recently, bla genes encoding resistance to β-lactams have frequently been identified in drinking water systems [14, 23, 52].
The antibiotic resistance levels in municipal wastewater can vary greatly because of its complicated chemical compositions and distinct microbial communities. As shown in Table 1, the ARB concentrations mostly ranged from 103 to 104 CFU/mL, and the bacterial resistance to tetracycline, sulfonamides, ciprofloxacin and ampicillin was commonly identified. Generally, the abundance of sulfonamide-resistant bacteria was higher than that of tetracycline-resistant bacteria in UWTPs [53]. The absolute abundance of investigated ARGs in municipal wastewater ranged from 103 to 108 copies/mL and the abundance of intI1 was 3–5 orders of magnitude higher than that in the sources of drinking water (Table 2), indicating that HGT can be greatly enhanced with the enriched microbial population. Hence, UWTPs have been regarded as important hotspots of antibiotic resistance in recent years [54] and more effective solutions to ensure the ecological safety of wastewater treatments are urgently needed.
3. Common Methods of ARB/ARGs Detection in Drinking Water/Waste Water Systems
Researchers have proposed a few qualitative and quantitative methods for the identification of ARB/ARGs in composite environmental media, which can be mainly classified into culture-dependent methods (CDM) and molecular methods. Several sources of database (Google Scholar, Engineering Village and Web of Science) were searched with relevant key words of antibiotic resistance and drinking water/wastewater, and fifty-four/forty research studies reporting the antibiotic resistance prevalence in drinking water/wastewater treatment systems in the last 10 years were collected. The ARB/ARG detection methods used in these investigations are enumerated in the venn diagram (Fig. 3). The results revealed that qPCR is frequently used in wastewater systems (72.5%) and drinking water systems (46.3%), confirming that qPCR plays a dominant role in the quantitative characterization of antibiotic resistance. 16S rRNA sequencing is commonly combined with CDM to identify the isolated bacteria. When compared with qPCR, the application of metagenomic next-generation sequencing is quite limited, accounting for only about 13.0% and 10.0% of all drinking water system and wastewater system studies, respectively.
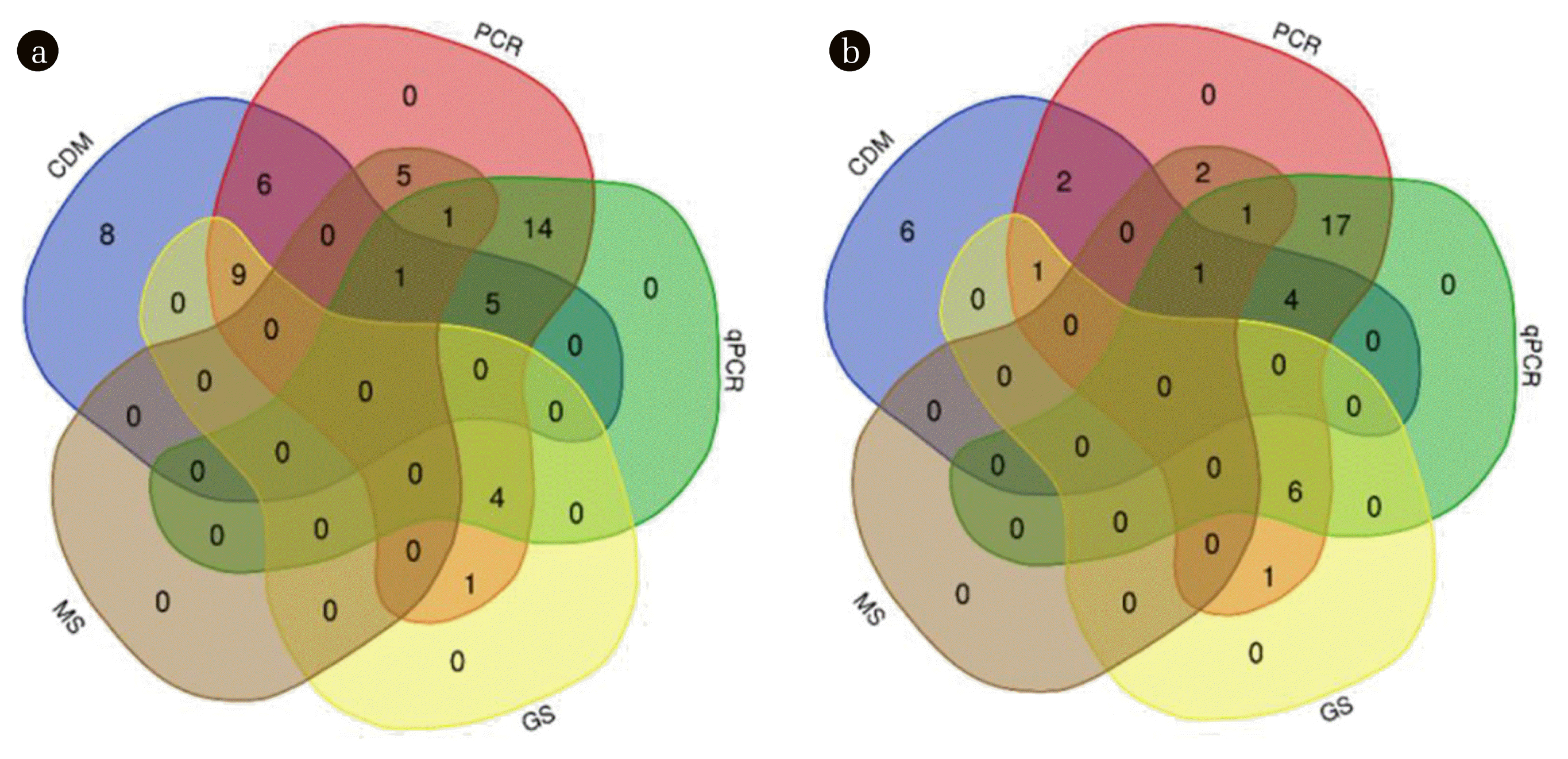
The combination of various detection methods applied in (a) the drinking water system (54 studies) and (b) the waste water system (40 studies) (CDM: culture-dependent method; GS: 16S rRNA gene sequence; MS: metagenomic next-generation sequencing) [6, 12–14, 17–19, 22, 23, 31, 32, 38, 52–54, 64, 66, 68, 69, 74, 76, 79, 80, 82–152].
For CDM, specific bacterial groups (indicator bacteria, pathogenic microorganisms or total culturable heterotrophs) are recovered from the filters. Subsequently the extracted bacteria will be cultured on the selective medium spiked with various antibiotics, and the antibiotic concentrations can be selected considering their resistance breakpoints based on the guidance of Clinical Laboratory Standards Institute (CLSI). The resistance/susceptibility profiles are then obtained based on the clinically standardized micro-dilution methods or disc diffusion methods. The micro-dilution methods determine the minimum concentration of specific antibiotic that completely inhibits the bacterial growth (i.e., MIC) to interpret the antibiotic susceptibility (sensitive, intermediate or resistant). Disc diffusion methods culture the bacteria with specific antibiotic-impregnated filter paper disks on the medium and determine the antibiotic susceptibility by bacterial growth or absence zone around disks. On this basis, the ARB counts and antibiotic-resistant ratio (dividing ARB counts by the total counts without antibiotics) can be calculated to evaluate the resistance loads of water environment. For example, Bergeron et al. [74] used Kirby-Bauer disk assay to define the resistance level of Enterobacter cloacae isolates in the source of drinking water and found high amounts of total and intermediate resistance against ampicillin. Although CDM are easy and convenient to operate, they can only identify the phenotypes of the viable and culturable indicators. However, more than 99% of the total bacteria cannot be cultured under laboratory conditions [75], causing the insufficiency of CDM in assessing the antibiotic resistance levels of environmental microorganisms. Additionally, some bacteria may be in a “viable but non-culturable” state after treatments, which can lead to false-positive results for the evaluation of ARB removal efficiency.
Recently, molecular methods have been extensively applied to gain insights into the emergence of antibiotic resistance at the genetic level [22, 69, 76]. Unlike CDM, culturing and enriching ARB is not required for molecular methods. Rather, genomic DNA for molecular detection is extracted from membrane filters by DNA spin kits according to the protocols. The DNA sections of interest are then amplified by polymerase chain reaction (PCR) and further quantitatively analyzed by real-time quantitative PCR (qPCR) with predefined primers. The advantages of qPCR mainly lie in: (i) saving time (2–3 h), (ii) detecting multi-gene, (iii) high accuracy (± 10%–20%) [71, 72], and (iv) high sensitivity. QPCR is known as the most sensitive and specific technique for the detection of nucleic acids. For instance, the traD gene qPCR was still able to detect in all the seven samples at 0.4 copy/μL [77]. Additionally, qPCR can be used to determine the abundance of iARGs, eARGs and MGEs through different pretreatments, enabling the prevalence and transmission of antibiotic resistance to be better characterized. However, only known genes can be identified by qPCR, implying that these methods would fail to estimate the overall resistance level to specific antibiotics [29]. Instead, researchers tended to assess the total abundance of several representative ARGs (e.g., using the total abundance of sul1 and sul2 to estimate the sulfonamide-resistance level). Furthermore, standardized operating instructions and integrated evaluating systems have not yet been established for qPCR; hence, the results from different studies are not highly comparable. As reported by Yoon et al. [78], the sensitivity of qPCR would be affected by the length of the amplified subsequences. In this study, the measured ARG (ampR and kanR) removal efficiency during chlorination and UV irradiation was relatively higher than that previously reported under comparable conditions [70, 79], perhaps because of the longer qPCR amplicons (800–850 bp) [78]. In recent years, the molecular methods have frequently been used in ARG detection with the popularization of more efficient and accurate qPCR technologies (such as high throughout qPCR). However, CDM cannot be completely neglected. Indeed, CDM are more appropriate for analyses relevant to ARB, such as determining the multidrug resistance ratio and examining the impact of trace-level antibiotics exerted on specific microbial communities [80].
Substantial ARB/ARGs can be identified by the above culture and molecular methods, but others cannot be detected due to limitations of the cultivability of bacteria and the types of predefined gene probes. To combat this problem, marker gene amplification metagenomics (e.g., 16S rRNA sequencing) and full shotgun metagenomics (e.g., metagenomic next-generation sequencing) are increasingly being adopted to unbiasedly collect all the genetic information in samples and obtain high resolution genomic analysis of the unculturable microbes. Additionally, the microbial composition of aqueous matrices can be synthetically analyzed (with special attention given to the dominant and pathogenic population). For instance, Lu et al. [22] evaluated the bacterial community diversity at the phylum level and genus level by 16S rRNA sequencing, and then studied the co-occurrence patterns between the investigated ARGs and the potential dominant hosts by network analysis. Moreover, alterations of functional antibiotic resistomes and the co-occurrence patterns of ARGs and MGEs (integrons, plasmids, and transposons) before and after disinfection in full-scale DWTPs were also assessed by metagenomic next-generation sequencing [17].
There are some differences in the selection of ARB/ARG detection methods between drinking water and wastewater treatment systems, which may be attributed to the distinct contents of bacterial population and masking impurities. As shown in Fig. 3, CDM are more frequently applied in the drinking water systems (53.7%) compared to that in wastewater systems (30%), demonstrating that CDM remain major techniques for the assessment of health-related issues in low impacted waters. Moreover, the pretreatment steps also vary between the water and wastewater treatment systems. For UWTPs, 10–500 mL wastewater samples should be pre-filtrated for available assays [69, 70], but increasing volume of membrane filtration (5–10 L) becomes essential for DWTPs on account of lower bacterial contents [80]. Recently, researchers tended to associate the overall antibiotic resistance level with several potential indicative ARB/ARGs in water environment. As illustrated in Table 1 and Table S4, the intestinal bacteria (e.g., Enterobacteriaceae and Enterococci isolates) were generally applied as indicators in CDM for both water and wastewater samples. Sul and tet genes have been suggested as reliable indicators for antibiotic resistance monitoring in the wastewater [29]. For example, He et al. [81] confirmed that sul1 and tetW were suitable to surrogate the anthropogenic ARG sources in boiler feedlots (correlations of sul1 and tetW to all ARGs: R=0.96 and 0.86, respectively, p < 0.01). Similarly, intI genes were also found significantly correlated with the investigated sul and bla genes (p < 0.01) in the treated landfill leachates and its effluent-receiving waters [86]. In the drinking water treatment systems, significant correlations between the relative abundance of ARGs and MGEs (e.g., intI genes and Tn916 transposon mentioned by Lu et al. [87]) have been highlighted, and MGEs are regarded as suitable indicators for the assessment of resistance transfer capacity. As reported by Hu et al. [4], sul1, tetC, strB and ermB were significantly correlated to the intI1, presumably implying that the residual ARGs were still capable of regeneration and further transmission.
In summary, CDM can only identify the antibiotic resistance phenotypes of specific microorganisms based on relevant clinical standards, and information about the unculturable bacteria is not available. Therefore, it is incapable for CDM to evaluate the antibiotic resistance levels of environmental microorganisms. This deficiency can be made up by qPCR technologies, which collect the genetic information from both culturable and unculturable bacteria. As demonstrated in Fig. 3, qPCR is extensively used for quantitative analyzing the ARGs in DWTPs and UWTPs nowadays. However, the measurable ranges of qPCR are limited by the known genes, in other words, information about the unclassified ARGs is difficult to obtain, causing the insufficiency of qPCR detection in assessing the overall resistance level of specific antibiotic. In this regard, comprehensive genetic materials can be collected and efficiently examined by the metagenomic next-generation sequencing. This method provides the most information about ARB (culturable or unculturable), ARGs (classified or unclassified) and co-occurrence patterns of ARB and ARGs. The metagenomic sequencing technologies have not been widely adopted due to capital and technology constraints for the present, while it is expected to be a prerequisite for evaluating the environmental resistomes in the future.
4. The Fate of Antibiotic Resistance in Full-scale Drinking Water/Wastewater Treatment Plants
To date, the antibiotic resistance removal effects and underlying mechanisms of typical water treatment technologies (e.g., oxidation and disinfection) have been systematically elucidated in depth based on the lab-scale experiments [78, 79, 149, 153]. However, information regarding the migration of ARB/ARGs in full-scale DWTPs and UWTPs (especially in the sequential processing units) is not adequate yet, resulting in a fragmented understanding of the resistance removal capacity of current treatment facilities. To monitor antibiotic resistance levels in engineering fields, more mechanism studies are needed.
4.1. Drinking Water Treatments
Physical processes (e.g., coagulation, sedimentation and filtration) are designed to remove suspended particles, after which disinfectants are applied to inactivate pathogenic microbes for conventional drinking water treatments. Advanced treatments (e.g., ozonation-biologically activated carbon (O3-BAC) process and membrane technology) are also used in some newly constructed DWTPs to further eliminate organic pollutants from water. In this section, the fate of antibiotic resistance during coagulation and sedimentation, filtration, chlorine disinfection and ozonation is discussed according to available studies. The resistance removal efficiency of individual processes was evaluated by analyzing the ARB ratio (incubated ARB counts divided by the heterotrophic plate counts) as well as the absolute or relative ARG abundance (the relative abundance was normalized by 16S rRNA) in the influent and effluent of specific utilities. Herein, seven studies determining the ARG abundance in full-scale DWTPs were selected from Web of science and Engineering Village with the key words of antibiotic resistance or antibiotic resistance genes and drinking water or drinking water treatment. Because of the low amount of culturable bacteria isolated from drinking water systems, it is difficult to quantitatively analyze the alterations of ARB among the sequential processing units [93], and in this regard only one such study was collected with the key words of antibiotic resistance or antibiotic resistant bacteria and drinking water or drinking water treatment. The removal efficiency of six common ARGs and 16S rRNA in the advanced water treatment (O3-BAC) process was determined to be a logarithmic reduction by Su et al. [64] (Fig. 4). On this basis, the effects of each processing unit on the fate of ARB/ARGs are summarized as follows.
4.1.1. Coagulation and sedimentation
General theories suggest that, during the flocculation process, partial bacteria and extracellular DNA will be condensed by electric-double-layer compression and charge neutralization with the assistance of positively charged coagulants (e.g., polyaluminium chloride and polyferric chloride), after which they are separated from water in the precipitation process. In addition, a small proportion of bacteria can be directly inactivated by ferric chloride since the ferric hydrate ions can oxidize intracellular functional enzymes after being absorbed by bacterial cells [154]. Therefore, the absolute abatement of the antibiotic resistance caused by coagulation and sedimentation is mainly credited to the condensation and partition of host bacteria as well as ARGs, and ferric hydrate ions may directly destroy ARB through oxidation.
In actual DWTPs, the ARB ratios toward ampicillin, kanamycin, rifampicin, chloramphenicol, and streptomycin (1 μg/mL) detected in the effluent of sedimentation tank were lower than 2×10−5 and did not significantly increase compared to the influent [80]. Moreover, as shown in Fig. 4, attenuation of ARGs was observed during coagulation and sedimentation treatments. Specifically, the removal efficiency of 27 typical ARGs mostly ranged from 0.5 log to 1.5 log (absolute abundance) when 32 mg/L polyaluminum chloride was used as the coagulant with higher removal of tetA and qnrA (4.57 and 3.46 log, respectively) [64]. Similarly, satisfactory removal of intI1, sul1, and strA genes was observed during the conventional flocculation and settlement process, except for the ermC gene, which was not removed and instead increased greatly [23]. The relative abundance of bla genes and some other efflux ARGs did not obviously increase [76], implying that these ARGs were not be selectively enriched in this process. The varied ARG removal during coagulation may have been a result of distinct removal of their host bacteria. For instance, previous studies revealed that there was less tendency for E. coli (Gram-negative) to attach to particles when compared with Staphylococcus sp. (Gram-positive) because of its less hydrophobic cell wall [155]. Moreover, the type and dosage of coagulants are considered to be potential influencing factors of antibiotic resistance removal efficiency. Li et al. [156] compared the ARG (sul1, sul2, tetO, tetW, tetQ and intI1) removal rates of two coagulants (ferric chloride and polyferric chloride) at different concentrations. Taking the removal of sul1 as an example, the removal efficiency increased as the concentration of polyferric chloride increased, peaking (2.9 log) at a dose of 12 mg/L. When ferric chloride was used, no significant correlation between the coagulant dose and sul1 removal efficiency was observed, and the maximum removal of 2.7 log was obtained at a dose of 18 mg/L. The different ARG removal efficiencies during coagulation with ferric chloride and polyferric chloride were attributed to the discrepant Fe species [156]. However, there is still a knowledge gap regarding the effects of other operating conditions on antibiotic resistance during flocculation (e.g., pH, water temperature, the addition of coagulant aids); therefore, further mechanism studies are warranted.
4.1.2. Filtration
The removal of antibiotic resistance during the filtration process can be credited to the interception or adhesion of ARB by filter media. Dullemont et al. [157] observed that the removal rate of E. coli in a slow filter reached 3.7 log during summer. It can be inferred that the filter is equipped to entrap a portion of ARB and thus reduce the total resistance. However, in this review, we failed to obtain an accurate decrease of ARB abundance during the filtration process in DWTPs. The ARB ratios did not obviously increase after filtration as detected by Bai et al. [80]. Perhaps the impact of sand filtration on ARB is so limited that it cannot be identified by CDM.
As shown in Fig. 4, limited reduction of sul1, sul2, tetA and tetW (less than 1 log) was observed during the sand filtration process [64]. Similarly, no notable removal of various ARGs was observed in other studies [76, 158]. Xu et al. [76] reported that the absolute abundance of sulfonamide-resistant and vancomycin-resistant genes was instead obviously amplified by this treatment, and the relative abundance of aminoglycoside-resistant and tetracycline-resistant genes also increased. Controversial results have also been obtained in studies of the effects on the absolute abundance of 16S rRNA, which is commonly used to identify bacteria at the genus level. For example, a reduction of 0.51 log 16S rRNA was detected by Su et al. [64] while a 0.1 log increase was noted by Hu et al. [23]. The discrepancy may have been a result of different sampling periods (e.g., before and after the backwash), which could have altered the absolute abundance of vancomycin- (32 types) and sulfonamide-resistant genes (sul1, sul2, sulA-folP-01, sulA-folP-03) on the biological active carbon as confirmed in another study [158]. In addition, the types of filter media can influence the fate of ARGs because of their distinct adsorption capacity [159]. For instance, a higher removal rate of culturable staphylococci (up to 0.7 log) was observed for a combined granular active carbon-sand media than a sand filter (up to 0.4 log) [160]. Other operating parameters (e.g., filtering velocity, column height and backwash cycle) may also influence ARG removal during the filtration process, but relevant studies were not revealed by our investigation.
4.1.3. Chlorine disinfection
Based on the available literature, chlorination was selected as the representative disinfection method for this section. Chlorine is advantageous for inactivation of bacteria because of its strong oxidizing power in neutral and acidic solution, which enables it to destroy cell membranes and affect multiple enzyme systems, releasing the intracellular materials (e.g., proteins, RNA and DNA). Accordingly, the removal of ARB and partial ARGs can be expected in response to chlorine treatment.
In a lab-scale chlorine disinfection experiment conducted in phosphate buffered solutions, the amount of ARB (E. coli with intracellular ampR gene) was effectively reduced by more than 3 log (calculated by plate counting) in response to the chlorine exposure at around 0.1 (mg×min)/L [78]. Additionally, an approximately 1 log reduction of ARB was observed in wastewater samples after the laboratory disinfection with 2 mg/L NaClO for 30 min, while the ARB ratio toward tetracycline increased from 3.4% to 16.7% at 8 mg/L chlorine [161].
However, limited attenuation or even proliferative effects of several ARGs (e.g., sul, tet and erm genes) as well as MGEs are generally identified for chlorination in full-scale DWTPs, and varying results have been observed. For example, approximately 2.0 log reduction and 1.5 log amplification of intI1 were identified (reported as absolute abundance) after chlorination during conventional and advanced treatment (O3-BAC) processes, respectively, even though the chlorine dosages were comparable [23]. Moreover, Xu et al. [76] found that the concentrations of substantial ARGs decreased 1–2 log after chlorine disinfection for both conventional and advanced treatment (O3-BAC) processes, whereas the relative abundance of various ARGs increased.
The abnormal increase in ARB ratios and ARG relative abundance after chlorination (at lower concentrations) indicates that the remaining microbes (which are mostly chlorine-tolerant, e.g., Pseudomonas and Acidovorax) are highly equipped with ARGs. This has been defined as a “co-selective” or “cross-resistant” effect by some researchers [113, 162]. Based on the current theory, the potential mechanisms of this “co-selective” effect are summarized as follows:
Simultaneous activation of antibiotic resistance caused by the self-adaptive process of bacteria to disinfectants. Bacteria can overcome such selective pressure problems by a series of SOS processes, such as altering the permeability of cell membranes, precluding the molecular transport and upregulating the efflux pump systems, which further facilitates the induction of antibiotic resistance [162]. For instance, Hou et al. [153] found that 25 efflux pump genes were upregulated after the treatment with 4 mg/L sodium hypochlorite for 20 min. There is some evidence that bacteria can undergo adaptive mutations under selective pressure [163], which may explain the significant increase (p < 0.05) in multidrug resistance genes after chlorination observed in another study [108].
Enhanced ARG transfer under oxidative pressure. A recent study revealed that subinhibitory exposure of chlorine (i.e., 0.1–1 mg/L Cl2 for free chlorine) caused augmented intragenera conjugative transfer compared with controls (3.4–6.4 fold) [164], confirming that HGT could be promoted by the low concentrations of chlorine used in actual DWTPs and water distribution system. The underlying mechanisms comprise cell membrane regulation, SOS response and reactive oxygen species formation induced by chlorine exposure [164]. Another study also confirmed that SOS response could alleviate the repression of SXT (an integrating conjugative element encoding resistance to chloramphenicol, sulfamethoxazole, trimethoprim and streptomycin [165]), thus enhancing the related antibiotic resistance transfer [166].
Theoretically, extracellular DNA can be oxidized by chlorine, but only a small portion of eARGs are damaged under actual treatment conditions because of the preferential consumption of chlorine by other cellular components and reductive contaminants. Yoon et al. [78] reported that the eARG (ampR and kanR) concentrations did not decrease and even increased slightly in response to a chlorine dose of lower than 12.5 mg/L (treated for 1 hour) in UWTP secondary effluent, while a 4 log reduction of eARGs was achieved in response to the chlorine exposure of ~50 (mg×min)/L in phosphate buffered solutions. Considering the insufficient oxidation capacity and “co-selective” effect of chlorine, improved technologies such as UV/chlorination systems have been extensively investigated for their capacity to remove ARGs [79, 149]. For instance, up to 4 log attenuation of the sul1 gene was achieved after the treatment with a UV/chlorination system for 10 min (UV intensity: 200 μW/cm2, free chlorine: 20 mg Cl2/L), which was more efficient than simple UV or chlorination treatments (about 3.5 log and 1.0 log sul1 removal, respectively) [149]. The authors believed the generation of more chloride radicals in this composite system was responsible for the enhanced ARG removal [149].
Even though the effectiveness of chlorination on the removal of ARB/ARGs has been widely investigated, the effects of various environmental factors (e.g., pH, organic matters, disinfection by-products) have seldom been discussed. Moreover, the mechanisms of antibiotic resistance attenuation (e.g., dynamic response, ARG expression, changes in HGT frequency) during disinfection processes should be investigated in-depth.
4.1.4. Ozonation
Ozone is an efficient and eco-friendly oxidant that is extensively applied for treatment in newly constructed waterworks through two approaches: pre-ozonation and post-ozonation. Pre-ozonation, which is regarded as an alternative to pre-chlorination, is applied to preliminarily oxidize pollutants and further intensify the organic removal that occurs during the coagulation and sedimentation process [167]. In post-ozonation process, ozone oxidation is typically combined with activated carbon filters, during which time the complex macromolecular organic materials can first be decomposed into small molecules by ozone, after which the biodegradable organic materials and some ammonia nitrogen are removed by biological degradation in the filters.
Similar to chlorine, ozone can oxidize the cellular components of ARB and directly destroy the structure of ARGs [78, 168]. General theories suggest that ozone inactivates bacteria through two prevalent pathways: (i) crossing the cell membrane of bacteria, then breaking down the internal glucose oxidase and destroying the nucleic acids; (ii) reacting with the outer membrane lipoproteins and lipopolysaccharides, causing cell membrane damage and culturability loss before affecting the intracellular nucleic acids. Zheng et al. [161] claimed that after ozonation (6 mg/L ozone for 10 min), most of the remaining intI1 presented in the form of cell-free DNA, indicating that intracellular intI1 was released from the investigated ARB (Bacillus subtilis) and mechanism (ii) mentioned above might be the main pathway in this case. However, Czekalski et al. [169] noticed that the membrane damage to E. coli during the ozonation process lagged behind the loss of cultivability, implying that the impact of mechanism (i) should not be excluded.
Studies of full-scale DWTPs have demonstrated that limited alteration of the ARB ratios was achieved by pre-ozone treatment [80] and the absolute abundance of ARGs would either slightly decrease or increase depending on gene types [18]. Interestingly, Hu et al. [23] noted that pre-ozone treatment could enhance the resistance removal effects of the subsequent coagulation and filtration process (analogous to the enhanced organic removal mentioned above). Compared to pre-ozonation, the post-ozonation process was more efficient in abatement of ARB and ARGs [18, 76, 80], probably because of the higher ozone doses and the presence of fewer ozone-depleting substances in the influent of post-ozonation. As demonstrated in Fig. 4, reductions of 0.38 and 0.46 log (reported as absolute abundance) were identified for sul2 and ermB during post-ozonation, respectively, comparable to the removal of sul1 and sul2 (up to 0.5 log) reported by Hu et al. [18]. In addition, the relative abundance of multiple ARGs also decreased [76]. However, there were some conflicting results. For example, tet genes were attenuated according to Xu et al. [76] while they were enriched (up to 3 log) in the present study (Fig. 4) [64]. The underlying mechanisms for these changes are still unclear.
Although biological activated carbon (BAC) filters can effectively degrade organic pollutants, recent studies have shown that this process failed to remove antibiotic resistance determinants or even enriched them, presumably due to the presence of favorable nutrients and sufficient hydraulic retention time (HRT) for ARB proliferation. For example, the ARB ratio toward kanamycin increased from 2×10−5 to 8×10−5 after being treated with a BAC filter [80]. A variety of ARGs (sul1, sul2, ermB, tetA, tetM, tetO, tetQ, tetS, tetW, tetX, cfr, cmlA, fexA, fexB, floR, oqxB, qeqA, qnrA, qnrB, qnrD and qnrS) were enriched by 0.03–5.12 log in the BAC effluent, and their relative abundance also dramatically increased [64, 76].
Some lab-scale or pilot-scale tests have attempted to elucidate the dose-dependent rules between the CT values (residual concentrations of the disinfectant multiplied by contact time) of ozone disinfection and the removal efficiency of antibiotic resistance. For phosphate buffer systems, He et al. [170] observed that ozone exhibited preferential reactivities toward GC bps and 5′-GT-3′ doublets in the given ARG (blt genes in Bacillus. subtilis strains) sequences. In this study, up to 3 log reduction of iARGs was achieved during ozonation at a low CT value of 0.4 mg/(L·min) [170]. For wastewater samples (secondary effluent from UWTPs), Czekalski et al. [169] noted that the most sensitive bacteria (about 1 log) could be initially inactivated (losing the culturability and cell membrane integrity) within 0.1 s by ozonation (2.5 mg/L), while according to the membrane integrity tests, the majority of autochthonous bacteria (approximately 103 CFU/mL) remained resistant at higher CT values (5 mg/L ozone, contact time > 30 min). The loss of sul1-stability only occurred when the ozone doses exceeded 1.89 mg/L, and the abundance of sul1 was abated by 2 log at an ozone dose of 2.15 mg/L. Similarly, Zheng et al. [161] found that although the concentrations of culturable bacteria were reduced by 1–2 log after ozonation (4 mg/L ozone applied for 10 min), the sulfamethoxazole-resistant ratio increased from 29.43% to 77.23%, implying that remaining ARB might be more tolerant to ozone. Moreover, the authors observed that tet and sul genes were reduced by 49.2% and 34.5%, respectively, at an ozone dose of 2 mg/L. The DNase I processing trials showed that ozonation caused cell membrane damage and release of substantial free DNA (containing ARGs) [161]. It can be inferred that ozonation efficiently removes the ARGs under ideal conditions, but is still limited by complex microbial communities and competitive or masking effects of other organics in full-scale treatment facilities.
The antibiotic resistance removal efficiency of ozonation is also related to pH values. Lower pH values (pH 6 and 7) were found to lead to higher removal efficiency of ARB (E. coli DH5α) and pB10 plasmids (containing ARGs) at an ozone dose of 3 mg/L [171]. The authors speculated that the maintained concentrations of ozone molecules was more likely responsible for the enhanced ARB and ARG removal [171]. Under alkaline conditions, ozone is partially converted to hydroxyl radicals, which are highly reactive and nonselective species [168]. However, marginal removal effects of hydroxyl radicals on the iARGs have been identified in recent studies [170, 171], which can be attributed to its consumption before reaching the intracellular materials. Moreover, the co-effects of ozone-depleting substances (such as humic acid and suspended solids) that might present in actual DWTPs/UWTPs were also examined [171]. The results revealed that the increase in suspended solids would hinder the removal of resistant E. coli DH5α, pB10 plasmids and pB10 plasmid transfer as expected, but the addition of humic acid (5 mg/L) promoted the ARB removal during ozonation. The underlying reasons for this were unclear [171].
Ozonation is generally considered a promising technology for removal of ARB and ARGs. Unlike chlorination, ozonation has been found to have less proliferative effects on the ARG relative abundance in full-sale DWTPs. However, it should be noted that the combined biological filters will selectively enrich the ARB, therefore, controlling strategies are urgently needed. Moreover, the removal effects of ozonation on the intracellular resistomes and corresponding influencing factors (e.g., ozone dose, pH and various organic interferents) warrant further investigations.
4.1.5. General evaluation
The antibiotic resistance levels can be efficiently reduced after the full-scale drinking water treatment processes. As reported by Bergeron et al. [104], ARB were only identified in the source water of a local DWTP, and the finished water did not contain any viable bacteria, indicating that these processing units were effective in ARB removal. Multidrug-resistant isolates were identified in the effluents of DWTPs according to Kinge et al. [172], while the percentages of multidrug-resistance decreased compared to the influents. In general, 1–4 log reductions of common ARGs (e.g., sul1, sul2, tetA and ermB) and MGEs (e.g., intI1) could be achieved [18, 64, 76], with limited alterations of the less ubiquitous ARGs (e.g., qepA, sul3 and qnrA) [23], which could be explained by their lower initial concentrations. Significant removal effects (> 7 log) on sul1 and sul2 were identified for advanced treatment processes (O3-BAC), and less than 100 copies/mL of these genes were detected in the finished water [64]. Similarly, the absolute abundance of sul2 in the final water was less than 5.04×100 copies/mL in another study, suggesting the effectiveness of DWTP treatments [22]. However, Hu et al. [23] identified higher levels of sul genes (103–104 copies/mL) in the DWTP effluents and the underlying reasons for these changes warrant further studies.
ARB and ARGs can be initially partitioned by the solid-liquid separation units (e.g., coagulation and sedimentation process). For the sand filtration process, ARG migration is a joint result of physical interception and microbial growth that depends on the operational status of filters. The antibiotic resistance levels are effectively attenuated by the disinfection or oxidation technologies (e.g., chlorination, pre-ozonation and post-ozonation processes). Meanwhile, the preliminary elimination of suspended solids (including attached bacteria) and organic materials during physicochemical treatments (coagulation, sedimentation and filtration) can further facilitate ARG removal in subsequent oxidation steps, as a result of the weakening of competitive or masking effects [149]. However, concerns have raised that the lower concentrations of chlorine used in DWTPs and water distribution system may lead to the increase of ARB ratios and ARG relative abundance, i.e., co-selective or cross-resistant effect, which poses a threat to the biosafety of drinking water. The enrichment of antibiotic resistance in BAC effluents has also attracted researchers’ attention. Augmented doses of disinfectants and combined disinfection processes (e.g., UV/chlorination mentioned above) are promising for enhancing the ARG removal in DWTPs, and the related dose-dependent rules need in-depth studies.
4.2. Wastewater Treatments
Wastewater treatment processes are classified into primary, secondary and tertiary treatment depending on the distinct target pollutants and removal mechanisms [173]. Special facilities (e.g., grids, grit chambers and pre-sinking tanks) have been adopted to separate undissolved suspended solids in the raw water for primary treatment. Additionally, diversified biological processes with different operating modes have been designed for secondary treatment to remove biodegradable materials and nutrients such as nitrogen and phosphorus (e.g., activated sludge process, oxidation ditch, anaerobic-anoxic-oxic process (A2O) and membrane bioreactor (MBR)). Tertiary treatment mainly removes residual minerals, refractory organics and pathogens that remain in the secondary effluents, utilizing physicochemical technologies such as flocculation, filtration, adsorption, ion exchange and chemical disinfection.
In UWTPs, HGT may be further enhanced during biological processes with microbial enrichment, increasing the environmental risks of the receiving waters. Therefore, the fate of antibiotic resistance in full-scale UWTPs has been extensively studied recently. Scientific investigations were searched from Web of Science, Engineering Village and Google Scholar with key words of antibiotic resistance or antibiotic resistant bacteria or antibiotic resistance genes and wastewater or wastewater treatment. For instance, Mao et al. [152] examined the distribution of tet, sul, qnr and erm genes in UWTPs. The concentrations of these ARGs as well as their removal efficiency during the sequential processing units are summarized in Fig. 5 (a) and Fig. 5 (b), respectively. On this basis, the fate of ARB/ARGs in full-scale UWTPs is summarized below, and the technologies (flocculation, sedimentation and chlorine disinfection) that have been discussed for drinking water treatments in section 4.1 are not be detailed here.

(a) The average concentrations and (b) log removal values of common ARGs (20 tet genes, 4 sul genes, 4 qnr genes and 2 erm genes) during each processing unit in a full-scale UWTP (RW: raw water; E: effluent; PCT: primary clarifier tank; AaT: anaerobic tank; AT: anoxic tank; AeT: aerated tank; SCT: secondary clarifier tank; DU: disinfection unit) [152].
4.2.1. Primary treatment
The antibiotic resistance determinants can be preliminarily removed with large suspended particles by physical interception and sedimentation during primary treatment [174]. However, in actual UWTPs, the ARB ratio to ampicillin even significantly increased (p < 0.05) from nearly 30% in the influent to 50% after primary settling at an ampicillin concentration of 20 mg/L [86]. The underlying mechanisms for the increase in the ARB ratios observed during primary treatment are not clear; however, it may have been a result of the ARB amplification caused by certain selective pressure during HRT without sufficient interventions.
As illustrated in Fig. 5, limited reductions (less than 0.2 log) of the investigated ARGs by the primary clarifier tank was observed [152], similar to the results reported by Wen et al. [71] for the fine screen, grit chamber and sedimentation tank, where all the ARG removal rates were lower than 0.1 log. Moreover, the concentration of mecA gene increased slightly (0.09–0.55 log) after being treated by the pre-sedimentation tank [175].
It should be noted that the precipitated ARGs could be further enriched in the primary sludge due to the favorable conditions for microbial growth. For instance, Bengtsson-Palme et al. [176] observed that the total ARG concentration (quantified by metagenomics) in the primary sludge increased by about 3 orders of magnitudes compared to that in the influent (from 101 to 104 copies/ mL), indicating that the corresponding settlement should be treated carefully to reduce the ecological risks posed by its being landfilled or reused for agricultural purposes. Similarly, it is important to know the abundance of ARB/ARGs in the coagulated sludge in DWTPs, however, there is still little information available regarding this respect.
4.2.2. Secondary treatment
Activated sludge processes and their modified procedures are the most commonly applied biological wastewater treatment technologies. Using this method, organics and nutrients can be efficiently degraded by the bacterial biomass suspension (i.e., activated sludge). Theoretically, resistant microbes will be absorbed and then removed from water by solid-liquid separation. As expected, ampicillin-resistant and vancomycin-resistant Enterococcus faecium were reduced in count of 1–2 log CFU/100mL after being treated by secondary treatment (activated sludge or biological trickling filter beds) [62]. Similarly, a reduction in the ratio of ampicillin-resistant bacteria (19% to 11%) was observed after the activated sludge process, which was characterized by high sludge retention time (SRT) and low F/M ratio (30 d and 0.18 kg COD/(kg·VSS), respectively) [177], while the ARB ratio toward streptomycin significantly increased (p < 0.05) in another study [86] and the underlying reasons were not clear. The full-scale cyclic activated sludge system (including the secondary clarifier) could achieve 1.00–1.75 log removal of the ARGs (tetA, tetO, tetW, sul1, sul2 and blaCTX-M) and intI1 as observed by Wen et al. [71]. However, a mass of resistance genes would inevitably be transferred to the settlement and further enriched in the biosolids. The absolute abundance of common ARGs in activated sludge is summarized in Table S5. High concentrations of intI1 (108–1012 copies/g, as described in Table S5) were detected in activated sludge, implying that the activated sludge became ideal shelter for antibiotic resistance transmission [71, 72].
A2O is a modified activated sludge process that is widely introduced for further removal of nitrogen and phosphorus through the alternative operations of anaerobic, anoxic and aerobic reactors [178]. Similar to the conventional activated sludge processes, the absolute abundance of various ARGs was effectively reduced by 1–2 log and their relative abundance also decreased after treatment of A2O and the secondary clarifier [67, 152]. Different ARG alteration patterns were observed in the A2O process (excluding secondary sedimentation) by Lee et al. [179]. Specifically, the relative amount of tet X increased significantly (463.9%), while that of ermB decreased by 87.0%, and there were limited alterations in the amounts of sul and bla genes. As reported by the authors, most of the ermB hosts were Gram-positive bacteria, which were presumably responsible for the ermB attenuation due to their poor survival in the activated sludge [179]. It can be inferred that the co-occurrence patterns of ARGs and specific host bacteria have an impact on the fate of antibiotic resistance during biological processes; however, the underlying mechanisms need to be investigated further.
Theoretically, the co-occurrence patterns of ARGs and specific ARB will change significantly under different dissolved-oxygen concentrations. The fate of antibiotic resistance determinants in the sequential anaerobic, anoxic and aerobic bioreactors has been specified and compared in recent studies, but there have been several contradictory conclusions possibly because of the distinct operating modes. Some studies found that the ARG concentrations were greater in aerated tanks than that in anaerobic and anoxic tanks, indicating that aerobic conditions were more favorable for ARB proliferation and ARG transfer [19, 72, 152]. Conversely, Zhang et al. [69] noticed that the relative abundance of various ARGs (tetM, tetG, tetX, sul1, sul2, ermB, ermF and mcr-1) would decrease as the oxygen concentration increased (i.e., anaerobic tank > anoxic tank > aerobic tank). Chen and Zhang [70] demonstrated that there were no obvious differences between the ARG concentrations in the anaerobic, anoxic and aerobic effluents.
As shown in Fig. 5(b), the investigated ARGs accumulated in all of the bioreactors, among which the largest amplification was achieved in the anaerobic tank. However, the anaerobic conditions appeared to be more conducive to ARB/ARG removal in other studies. Wang et al. [72] emphasized the substantial ARG enrichment in the aerated tanks with limited amplification yielded in the anoxic and anaerobic tanks. Lamba and Ahammad [180] observed that a lab-scale anaerobic flow-through reactor was less likely to induce multidrug resistance genes than an aerobic process and Christgen et al. [106] confirmed higher resistance attenuation of a combined anaerobic-aerobic treatment process compared to a single oxidation process.
The resistance removal efficiency of the A2O process can be influenced by typical operating parameters (e.g., SRT, HRT and sludge load). Studies have confirmed the important role of SRT in ARG attenuation, which can be elucidated by its shaping effects on the activated sludge microbes. For instance, the absolute abundance of total ARGs decreased by 0.33 log while their relative abundance increased by 2.18 times with the extension of SRT (from 25 d to 50 d) as observed by Zhang et al. [69]. In their study, enrichment of intI1 in the water and sludge was also reported, indicating that longer SRT could promote HGT.
An oxidation ditch is a modified activated sludge process that utilizes longer SRT to remove the biodegradable organics [181]. Delayed aeration and continuous flow are adopted and preliminary settling is unnecessary in this system. McConnell et al. [67] reported that E. coli was effectively reduced by around 2 log after the treatment of an oxidation ditch. In this study, the absolute and relative abundance of various ARGs also decreased in aerated lagoons, and the total ARGs were substantially reduced by 1.36 log in subsequent secondary clarifiers.
MBR systems use the membrane technology as a treatment step with no return of active sludge to the biological reactor. Similar to the above biological processes, ARB/ARGs are absorbed by the activated biomass. Additionally, the attached ARB/ARGs can be directly intercepted in the first stage during membrane separation. Moreover, studies have shown that membrane fouling substances (e.g., extracellular polymers) would gradually diminish the pore size during filtration, thereby strengthening the capture of free ARB and ARGs [182]. Notably, although efficient removal of bacteria such as E. coli by MBR system has been widely verified [183, 184], the ARB removal of MBR has seldom been reported to date.
Various subtypes of ARGs were removed with reductions of 1–2 log by the full-scale MBR process, which was more effective than the other investigated biological processes (aeration tanks and cyclic activated sludge system) as noted by Wang et al. [72]. Similarly, Li et al. [185] highlighted the maximum ARG removal efficiency achieved by MBR, which was attributed to the enhanced solid-liquid separation of MBR. Overall, studies conducted to date indicate that MBR technologies are promising for the antibiotic resistance removal. However, some pathogenic bacteria have been shown to be selectively retained by the membranes [185], perhaps due to differences in the bacterial size and deformability, suggesting that the biosafety of MBR effluents need to be further evaluated.
In summary, sewage biological treatment processes mainly transfer attached ARB and ARGs to the activated biosolids. From this perspective, methods that can optimize the efficiency of solid-liquid separation will strengthen antibiotic resistance removal. It is commonly recognized that substantial ARB and ARGs can be partitioned from water by biological treatments (including the secondary clarifier), and this has been found to be more efficient than disinfection processes [67, 71]. Moreover, no selective amplification of specific ARGs has been identified in secondary effluents to date. However, as discussed above, ARB and ARGs are significantly enriched in activated biomass (Table S5), where complex pollutants (e.g., heavy metals) can contribute to further co-selection of antibiotic resistance phenotypes [69, 152]. Therefore, post-control of the accumulated resistance in sludge is needed to avoid extended pollution in receiving waters or soils.
The effects of SRT on the fate of antibiotic resistance during biological processes have been focused in relevant studies, and high SRT seemed more conducive to ARG removal. However, Turolla et al. [177] specified that two activated sludge processes with identical SRT achieved different ARB removal rates, indicating that the fate of antibiotic resistance was also affected by other factors (e.g., intricate chemical components, different initial ARB concentrations and microbial community compositions). These findings emphasized the need for caution when comparing the resistance removal efficiency of secondary treatment in full-scale UWTPs.
4.2.3. Tertiary treatment
Additional steps have been adopted after secondary treatment in full-scale UWTPs to further remove the bio-refractory organics, turbidity, nitrogen, phosphorus, metals, and pathogens. Some physicochemical (e.g., flocculation and filtration) and disinfection processes (e.g., chlorination and UV irradiation) are also involved in the tertiary treatment.
Flocculation is considered a promising process for ARG removal in full-scale UWTPs. For example, Lee et al. [179] found that 48.2%, 75.3% and 44.3% of the tet, sul, and bla genes were removed, respectively, during flocculation (using 62.4 mg/L polyaluminum chloride as coagulant). However, the opposite was observed in another UWTP investigated in the same study, where the abundance of sul and bla genes increased by 35.6% and 152.1%, respectively, and the lower coagulant dose (33.3 mg/L) was responsible for this discrepancy, although the dose-dependent mechanisms of ARG inactivation were not revealed [179]. In summary, the pre-settling tank, secondary settling tank and flocculating settling tank all remove pollutants from water through solid-liquid separation, but their resistance removal efficiencies are distinct. The adsorption capacity of activated sludge and flocs generated during secondary treatment and flocculation is stronger than that of inorganic particulates treated in pre-sedimentation, therefore, the former two processes perform better when applied for settling of ARB and ARGs.
Sand filtration has limited impacts on the fate of antibiotic resistance in UWTPs. As examined by Petrovich et al. [62], sand filtration or lagooning processes achieved 0.8 and 0.7 log CFU/100mL removal of ampicillin-resistant bacteria and vancomycin-resistant bacteria, respectively. McConnell et al. [67] also found that only a small portion of E. coli (less than 1 log) was removed during sand filtration. Moreover, the abundance of total ARGs (intI1, qnrS, sul1, sul2, tetO, ermB, blaTEM, blaCTX-M and mecA) decreased with an average reduction of 0.26 log during sand filtration [67]. Similarly, a series of pilot tests demonstrated that sand filtration could only reduce the AdvIPC (an antibiotic resistance plasmid) by 0.9 log, while the authors identified a synergistic effect between pre-filtration and chlorination that led to an additional removal (> 1.8 log) compared to samples that had been subjected to single chlorination treatment [73].
Chlorine and UV were selected as typical disinfectants in full-scale UWTPs based on the available literature. Chlorination (residual chlorine 0.2 mg/L for 30 min) achieved a 0.65–0.89 log CFU/mL reduction of norfloxacin-resistant bacteria in an Indian UWTP as reported by Kurasam et al. [186]. Lab-scale disinfection experiments showed that the counts of ARB further decreased as the chlorine dosage increased (during treatment for 30 min) and all of the ARB lost their culturability at 32 mg/L [161]. The authors emphasized the increase in the tetracycline-resistant ratio after being treated with 8 mg/L chlorine [161], while Turolla et al. [177] found that the ampicillin-resistant ratio in E. coli remained unchanged after chlorination (average NaClO dose: 1.8 mg/L, during treatment for 35 min), implying that chlorine did not act selectively on these ARB. For ARGs, the absolute abundance of sul genes decreased slightly by 0.37 log after chlorination (5 mg/L for 30 min) as shown in Fig. 5. Herein, the chlorine was found to be preferentially consumed by other reductive contaminants in the wastewater and converted to the less effective chloramine [79] before degrading the ARGs, resulting in the lower ARG removal rates. Moreover, during the lab-scale chlorination processes, the removal efficiency of sul1 increased from 1.39 log to 3.11 log as the chorine doses increased (from 5 mg/L to 25 mg/L, during treatment for 30 min), suggesting that augmented chlorine doses were required for efficient ARG attenuation in the UWTPs.
UV disinfection inactivates microorganisms by physical processes (e.g., light absorption), which primarily targets nucleic acid and subsequently leads to reduced cell culturability [27, 187]. Unlike chlorine disinfection, UV can penetrate the UV-transparent structures and cause damage to iARGs before cell membrane breakage [27, 78]. According to Zheng et al. [161], the amounts of tetracycline-resistant bacteria and sulfamethoxazole-resistant bacteria were reduced by 0.73 and 0.86 orders of magnitude after laboratory UV disinfection (UV fluence 10 mJ/cm2 for 30 min), respectively, and decreased to below 1 CFU/mL at an UV fluence of 80 mJ/cm2, which was comparable to the practical doses in UWTPs. The authors also emphasized that the investigated ARB had a greater tolerance to UV irradiation than non-ARB [161], while the opposite results were obtained by Turolla et al. [177], who found the ampicillin-resistant bacteria were selectively abated by UV disinfection.
The ARG removal rates of UV in full-scale UWTPs were found to be comparable to those of chlorination based on relevant studies. For instance, the absolute abundance of ARGs (tetA, tetO, tetW, sul1, sul2, blaCTX-M and intI1) was reduced by 0.20–0.33 log after UV disinfection [71]. Similarly, the average removal rate of total ARGs reached 0.56 log at an UV fluence of 250 mJ/cm2 [67]. However, the total ARGs increased 0.2 log after the UV disinfection with lower ultraviolet intensity (50 mJ/cm2) in another UWTP [67], suggesting that low-intensity UV might be insufficient to remove ARGs because of the UV screening effects of intricate components in wastewater.
4.2.4. General evaluation
Overall, ARB counts can be effectively reduced after being treated by full-scale wastewater treatments. For instance, average counts of 2.7 and 2.5 log CFU/100 mL for ampicillin-resistant and vancomycin- resistant Enterococcus faecium were removed from UWTP effluents compared to the untreated samples, respectively [62]. However, recent studies stressed that some ARB could not be completely inactivated. Occasionally, high counts of resistant strains (101–103 CFU/mL) were identified in the finished water, which were subsequently discharged into the receiving waters [55, 59, 186]. The changing modes of ARB ratio varied with antibiotic types, for example, the tetracycline-resistant ratio in E. coli decreased in UWTP effluents compared to raw water, while the amoxicillin-resistant ratio increased slightly as observed by Lefkowitz and Duran [86]. The multidrug resistance in E. coli also increased in the same study (especially for the resistance to more than four antibiotics) [86].
Common ARGs (e.g., sul, tet, erm, bla genes) were reduced by 1–3 log (reported as absolute abundance) in the investigated UWTPs. The relative abundance of sul1, ermB and intI1 significantly decreased (p < 0.05), while significant increase (p < 0.05) was identified for blaTEM, blaCTX-M and qnrS [68]. As demonstrated in Table S5, the ARGs were significantly enriched in activated sludge. Specifically, the absolute abundance of sul, tet, erm, bla and intI1 genes in sludge was 3 to 5 orders of magnitude higher than that in UWTP effluents. In view of the fact that ARB and ARGs are simply transferred from water to the activated biosolids without permanent inactivation, efficient disposal technologies are required to ensure the ecological safety of sludge reusing. After being treated by full-scale UWTPs, high concentrations of sul (> 105 copies/mL), erm (> 104 copies/mL) and intI1 (> 106 copies/mL) genes were detected in the effluents (Table S5), comparable or occasionally higher than the resistance levels in surface water according to Table 2, implying that the UWTPs are important hotspots of antibiotic resistance nowadays.
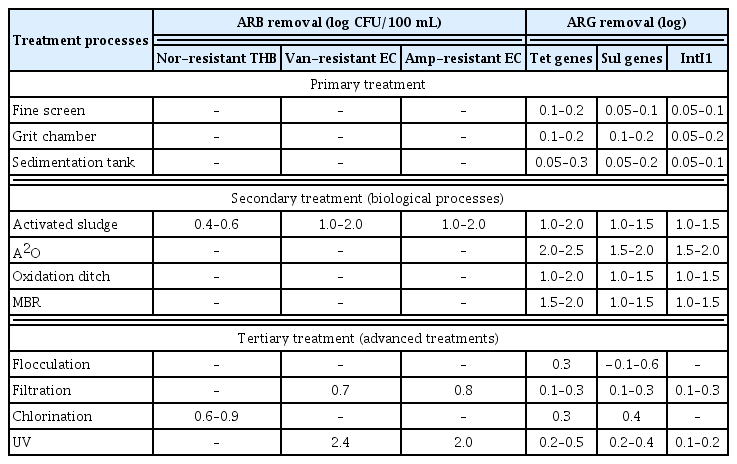
The ARB and ARG removal efficiency during the sequential processing units in full-scale UWTPs was estimated in Table 3 based on the above discussions. In general, the primary treatment has limited impact (< 0.1 log ARG removal) on the antibiotic resistance. The biological processes (including secondary clarifiers) generally exhibit the most significant removal effects on the ARGs (1.0–2.0 log), and the disinfection processes (UV and chlorination) take second place (0.1–0.5 log). Although the total abundance of ARB and ARGs can be substantially attenuated after wastewater treatments, the high resistance levels in finished water still cause concern. To mitigate the environmental risks, increasing doses of chlorine (or higher UV intensity) and combined disinfection processes can be adopted by UWTPs to further enhance the attenuation of ARB and ARGs. Moreover, it may be feasible to transfer most of the ARB and ARGs into activated biomass during biological processes and subsequently treat the sludge with efficient disposal technologies (e.g., incineration and digestion) to eliminate the antibiotic resistance.
5. Conclusions
This review has highlighted the prevalence and widespread transmission of antibiotic resistance determinants in various aquatic compartments (surface water, municipal wastewater, and drinking water), which poses a significant threat to human health. Therefore, it is important to understand whether the current water/wastewater treatment facilities are capable of ensuring the production of finished water with an acceptable level of antibiotic resistance. This would allow us to evaluate the human accessibility and environmental risks associated with these emerging pollutants. After being treated by the full-scale DWTPs, both ARB and ARGs can be effectively removed. Solid-liquid separation (e.g., coagulation and sedimentation), disinfection, and oxidation (e.g., chlorination and ozonation) play a dominant role in this process. For the UWTPs, ARB and ARGs can also be substantially reduced by the biological processes and advanced treatments. However, ARB and ARGs cannot be completely eliminated by the current treatment systems, and the residual resistance in treated water has raised concern. Moreover, there are still a few issues that should be paid reasonable attention. For instance, lower oxidative pressure exerted by chlorine or ozone results in the accumulation of ARB ratio and ARG relative abundance. Increased doses of disinfectants or oxidants and combined disinfection technologies can be used to solve these problems, but other effective strategies (e.g., membrane separation) need to be developed. All of these points require further understanding of (i) the impacts of the key operating parameters in each processing unit, (ii) co-selection mechanisms under low oxidative pressure, and (iii) dynamic mechanisms of ARB/ARG inactivation.
Supplementary Information
Acknowledgements
This study was supported by the National Key R&D Program of China (2019YFC0408801).
Notes
Author Contributions
R.G. (Ph.D. student) summarized the relevant literature and wrote the manuscript. M. S. (Professor) revised the manuscript.