AbstractIn this study, the mechanism of quorum sensing inhibition (QSI) by plant-oriented organic molecules, vanillin, was investigated. During the biofouling experiment with Pseudomonas aeruginosa as a model bacterium in the forward osmosis (FO) membrane process, the addition of 1 mM vanillin showed remarkable mitigation of biofouling without both bactericidal and bacteriostatic effects. Subsequently, bioassay of quorum sensing (QS) molecules and RNA sequencing of P. aeruginosa were conducted to figure out QSI mechanism. Interestingly, the bioassay result showed that the addition of 1 mM vanillin as the QSI was not sufficient to degrade the QS signal molecules such as N-Hexanoyl-DL-homoserine lactone. On the other hands, the analysis of RNA gene expressions exhibited that the significant down-regulation of QS receptors while there was not any noticeable changes in gene expressions related to synthesis of QS signal molecules. The down-regulation of QS receptor genes suppressed the hierarchical and interconnected QS system of P. aeruginosa, thus contributed as the main mechanism of QSI. The result of this study implies the potential applicability of QSI by plant-oriented organic molecules to mitigate biofouling in membrane processes where the concentration of organic matters is not important.
1. IntroductionWater reuse and desalination have been considered as effective ways to solve the global water scarcity [1]. In particular, membrane processes including nanofiltration (NF), reverse osmosis (RO) and forward osmosis (FO) have been widely applied in both desalination and water reuse processes as core technologies [2]. However, the biggest challenge facing membrane processes is biofouling caused by the consortium of organic matters, particulates and microorganisms which develop adhesive biofilm matrix by the release of sticky extracellular polymeric substances (EPS) [3]. Biofouling occurring in membrane processes causes the significant decline in the flux and quality of permeate [4].
As a method to control biofouling, physicochemical cleaning methods have been widely applied. However, physical cleaning with increase in the cross flow velocity has been shown to be ineffective in preventing or removing irreversible biofouling [5]. On the other hand, long term usage of chemical cleaning agents was proven to hamper the structure of the membrane [6]. Thus, the novel biological strategy, which can effectively hinder the build-up of biofouling without any damage on the structure of the membrane, is strongly required.
Bacteria are known to communicate with each other through quorum sensing (QS) by producing signal molecules and its biding with signal molecule receptors during the formation of biofilm [7]. The quorum sensing inhibition (QSI) has been studied in membrane-based water treatment systems to mitigate biofouling and three typical QSI mechanisms are proposed; suppression of signal molecules synthesis, degradation of signal molecules, and interference of signal molecules binding to the receptors [8, 9]. For instance, enzymes such as acylase and lactonase have been reported to degrade signal molecules, thus the enzyme-producing bacteria have been encapsulated in the beads, hollow cylinder, sheets or core-shell structure to control membrane biofouling in membrane bioreactor (MBR) [9–12]. However, economic feasibility and difficulties in maintaining enzymatic QSI activity are still hurdles for wide application of enzymes or enzyme-producing bacteria. Recently, plant-oriented organic molecules such as cinnamaldehyde, vanillin, and zingerone have been gained interest as one of sustainable alternative to enzymatic QSI [13]. These plant-oriented organic molecules are known to have the antibacterial activity as well as effectively mitigate the formation of biofilm at the concentration even lower than growth inhibitory concentration by the QSI activity [14–16]. Among these plant-oriented organic molecules, vanillin is a promising candidate to apply due to its nontoxic nature to human health, easy accessibility and ability to control biofouling in membrane-based water treatment processes [16]. In the previous study, when vanillin was applied RO process, it effectively mitigated biofouling and by the decrease in the amount of extra-cellular polymeric substances [17].
In our previous study with FO process, it was shown that the addition of vanillin retarded the formation of biofilm on the membrane surface [18]. Pseudomonas aeruginosa had been utilized as a model bacterium for biofouling experiment because the QS signaling system of P. aeruginosa included two AHL-based and two qui-none-based QS system [19]. Although three QSI mechanisms were proposed in our study, the detailed QSI mechanism of vanillin was not clearly elucidated [18]. In the previous study, inhibition of Las and Rhl genes of P. aeruginosa were shown to be tightly related to the biofilm formation [20]. Moreover, it is already reported on the genes encoding signal molecule synthesis and receptor protein [21]. Therefore, the expression of QS-related genes is essential to elucidate the mechanism of QSI molecules such as vanillin. Recently, RNA sequencing (RNA-Seq) has provided more precise quantification of transcriptomes [22]. The technology has been regarded as powerful tool to enable the improvement of functional genomics analysis including quantitative gene expression studies for the measurement of variability in expression level [23].
The objective of this study was to elucidate the QSI mechanism of vanillin among three proposed mechanism by the bactericidal and bacteriostatic experiments and by the comprehensive RNA sequencing analysis.
2. Material and Methods2.1. Bacteriostatic and Bactericidal Effects of VanillinThe experimental procedure to elucidate the major biofilm mitigation mechanism of vanillin is presented in the Fig. 1. The test for bacteriostatic and bactericidal effect of vanillin was designed as reported in the previous study [18]. Briefly, vanillin stock solution (Sigma Aldrich, USA) was prepared by dissolving vanillin in a DI solution with the final concentration of 10 mM. P. aeruginosa PAO1 was selected as a model bacterium throughout this study. For test of bacteriostatic and bactericidal effect of vanillin, 107/mL of cells were cultured at the presence of 0 mM, 0.1 mM, 1 mM and 10 mM of vanillin for 24 h. Potassium phosphate buffer (pH = 7.4) was utilized for dilution of the bacteria and vanillin. Bacteriostatic effect of vanillin was observed by measuring the optical density at 16 h and 24 h after the initial inoculation. For the bactericidal effect, samples were collected after 12 h and the Live/Dead tests of bacteria cells were performed with the SYTO9 and propidium iodide (Invitrogen, USA). Fluorescence images were taken in each sample and the live and dead cell was counted and averaged using the Image J program.
2.2. Inhibition of Biofilm Formation by VanillinThe thin film composite polyamide FO membrane (Toray, Japan) was stored at 4°C in DI water and sized 20.25 cm2 to be attached on the glass plate facing the active layer to the solution. The solutions containing 107 cells/mL of P. aeruginosa PAO1 were shaken with and without 1 mM vanillin for 24 h and 48 h at 25°C. Biofilm formed on the membrane was collected using silicon knife and filtered through 0.2 μm paper filter. The membrane was then dried at 105°C for 2 h. The difference in weight before and after the filtration was defined as the biomass.
2.3. Liquid Phase Bioassay for Determining The Degradation of Signal MoleculesDegradation of bacterial signal molecule by vanillin, the experiment with Chromobacterium violaceum CV026 was conducted followed by the method in our previous study [24]. N-Hexanoyl-DL-homoserine lactone (HHL; Sigma-Aldrich, USA) was used as an QS marker in this experiment. Briefly, HHL stock was solubilized in solution with 0.1% acetic acid and 100 mM ethyl acetate. HHL was then diluted with 10 mM potassium phosphate buffer to be concentration of 1 μM. The AHL indicator bacterium, C. violaceum CV026, was cultivated in Luria-Bertani (LB) medium for 24 h, centrifuged, and then rinsed three times with 0.9% NaCl. For the evaluation of QSI activity, 1 mM vanillin was supplemented in to the C. violaceum CV026 culture, and violacein was extracted by a protocol described elsewhere [25]. Finally, the supernatant in which violacien was contained was extracted, and the absorbance was measured using a UV spectrophotometer (Hach, USA) at 585 nm.
2.4. RNA Sequencing Library Construction and Sequence AnalysisThe preparation and analysis of RNA sequencing was performed according to the previously proposed method [26]. Briefly, P. aeruginosa PAO1 was cultured at 37°C in Luria-Bertani (LB) medium for 24 h and harvested. Then, P. aeruginosa PAO1 (108/mL) was cultivated again in LB medium for 48 h at the presence and absence of 1 mM vanillin. For the extraction of RNA, samples were centrigufed for 5 min at 13,000 rpm and washed three times with 0.9% NaCl. Samples were then purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and subjected to quantity and quality assessment by the BioAnalyzer 2100 using a DNA 1000 chip (Agilent Technologies, Santa Clara, CA, USA). Prior to cDNA library construction, RNA samples were enriched for transcripts using mRNA Purification Kit (Agilent Technologies, Santa Clara, CA, USA). To construct Illumina-compatible libraries, a TruSeq RNA library preparation kit (Illumina, San Diego, CA, USA) was used according to the manufacturer’s instructions. TruSeq RNA libraries constructed by PCR amplification were quantified using quantitative PCR (qPCR) according to the qPCR Quantification Protocol Guide, and the quality was assessed electrophoretically (Bioanalyzer 2100; Agilent Technologies). Paired-end type of read sequenced from the HiSeqTM 4000 platform (Illumina) was used and the reference genome was acquied from the control sample [27]. For the differentially expressed genes (DEG) analysis, expression values were estimated as reads per kilobase of the transcript per million mapped reads (RPKM) and normalized by totals. Subsequently, low quality genes were filtered and quantile normalization was conducted after the logarithm transformation [28].
3. Results and Discussion3.1. Bacteriostatic and Bactericidal Effects of VanillinTo investigate the quorum sensing inhibition of vanillin, bactericidal and bacteriostatic effects should be excluded. The growth curves of P. aeruginosa in LB supplemented with various concentrations of vanillin are shown in Fig. 2 (a). The inhibitory effect of vanillin on bacterial growth was negligible up to 1 mM (p-value > 0.05). However, the bacterial growth was retarded moderately at 10 mM of vanillin concentration (p-value < 0.05). In detail, the absorbance at 600 nm dropped to 1.68, which was 73% of control (0 mM) at 24 h, indicating that bacteriostatic effect occurred. In order to assess the bactericidal effect of vanillin, the portion of dead cells in each sample was observed and calculated by dividing number of dead cells with total cells as shown in Fig. 2(b). The portions of dead cells had no statistical difference in the presence of 0.1 mM (6.6%) and 1 mM vanillin (7.1%) compared to the absence of vanillin (8.7%) (p-value > 0.05), while, it increased significantly to 25.1% at the 10 mM vanillin. It indicated that the addition of 0.1 mM and 1 mM vanillin had no bactericidal effect to P. aeruginosa. It is in accordance with the previous study that the bactericidal effect was only shown at the high concentration of vanillin (15 mM ~ 30 mM) [29]. Thus, bacteriostatic and bactericidal effects were not observed at the concentration of vanillin up to 1 mM, and therefore, further experiments were conducted at 1 mM of vanillin to elucidate the QSI mechanisms of vanillin.
3.2. Applicability of Vanillin for the Biofilm MitigationThe amount of biofilm formed on the membrane in the presence of 1 mM of vanillin was only accounted to 38% and 45% of the mass compared to that of control (0 mM) at 24 h and 48 h, respectively (Fig. 3 (a)), and it is in agreement with our previous report that the decline of flux was retarded at the presence of 1 mM vanillin in the feed solution of continuously operated forward osmosis process [18]. From the result, the addition of 1 mM vanillin was effective in mitigating the biofilm formation on the membrane by QSI effect, not by bacteriostatic nor bactericidal effects.
3.3. QSI Mechanism of VanillinAs previously discussed, three QSI mechanisms have been proposed; the degradation of signal molecules, the suppression of signal molecule synthesis, and interference of signal molecules’ binding to the receptors. To examine the first mechanism, liquid phase bioassay using C. violaceum CV026 was conducted to test whether vanillin could degrade signal molecules. For the investigation of second and third mechanisms, the expression of functional genes responsible to the synthesis of signal molecules and receptors were analyzed by RNA sequencing.
3.3.1. Degradation of signal molecules by vanillinThe liquid phase bioassay using C. violaceum CV026 in the presence and absence of 1 mM vanillin was performed to verify the degradation of typical AHL-based signal molecule, N-hexanoyl homoserine lactone (HHL), by 1 mM vanillin. As shown in Fig. 4, there was not any noticeable changes in the concentration of HHL at the presence of 1 mM vanillin during the liquid phase bioassay. Therefore, the concentration of 1 mM vanillin could not degrade AHL. Consequently, it can be concluded that the mitigation of biofilm formation on the FO membrane was not contributed by the degradation of signal molecule by vanillin.
3.3.2. Suppression of functional genesThe QS signaling systems of P. aeruginosa includes two AHL-based (Las and Rhl genes) and two quinone-based (Iqs and Pqs genes) QS system. In each QS system, it is consisted of signal molecule synthesis (I followed by name of the genes, e.g. LasI) and receptor (R followed by name of the genes, e.g. LasR) protein, and there are several genes encoding these proteins [21]. After data pre-processing and quality check of raw data (5,703 genes) collected by transcriptome resequencing, 5,441 high-quality genes with 6,264,361 reads in two samples were obtained. Among collected transcriptome sequencing data, genes encoding QS system related proteins were matched with the data deposited in NCBI [30]. In Fig. 5 (a), the relative transcriptional expression of genes regarding AHL-based QS system (Las and Rhl) in the presence and absence of vanillin are presented. As shown in Fig. 5 (a), LasI and LasR genes were down-regulated 28% and 26%, respectively, in the presence of vanillin. Besides, expression of RhlI and RhlR genes were down-regulated 2% and 39%, respectively, in the presence of vanillin. Both synthesis and reception of signal molecules in Las and Rhl QS systems were down-regulated at the presence of vanillin. The decrease in the transcription level of AHL-based QS related genes is in agreement with the previous reports that the synthesis of N-Hexanoyl-L-homoserine lactone (C6-HSL) was shown to be reduced at the concentration of 1.7 mM vanillin [16]. The inhibition of synthesis and reception processes as well as the decrease in biofilm formation have been also reported from other studies. The meta-bromo-thiolactone (mBTL) also inhibited the expression of two quorum-sensing receptors of P. aeruginosa, LasR and RhlR, and led to the decrease in biofilm formation [20]. Thus, the down-regulation of LasI, LasR and RhlR genes was the one of QSI mechanisms of vanillin mitigating biofilm formation on the FO membrane surface.
In Fig. 5 (b), relative expressions of genes regarding quinon-based QS system (Iqs and Pqs) at the presence and absence of 1 mM vanillin are presented. From the result, AmbB and AmbC genes were 10% and 18% down-regulated, while AmbD and AmbE genes were 4% and 2% up-regulated by the addition of vanillin. PqsA, PqsB, PqsC and PqsD genes were 11%, 40%, 18% and 31% up-regulated by vanillin, respectively. Meanwhile, PqsR gene was found to be 18% down-regulated. Expression of Iqs and Pqs genes were inconsistent in the presence of vanillin with the concentration of 1 mM. Until now, the relation between the expression of Iqs QS system and biofilm formation is not clear. However, competitive antagonists of PqsR also reduced the virulence factor of the pyocyanin production by 74% during the addition of vanillin in P. aeruginosa PA14 [31]. Although there is no direct evidence on the effect of PqsR interference to the mitigation of biofilm formation, the formation of biofilm would also be reduced by the interference of PqsR.
P. aeruginosa is known to have a multi-layered hierarchy QS system consisting of four interrelated signaling mechanisms, and therefore, signal molecule and receptor complex of ruling hierarchy triggered the expression of lower hierarchy [32]. For instance, Las system was proven to govern the expression of both Pqs and Rhl systems, thus Las system was often described as being at the top of the QS hierarchy [32]. Thus, the binding of N-(3-oxodecanoyl)-homoserine lactone (OdDHL) with LasR controls the activation of both Pqs and Rhl QS systems. It is also shown that the disruption of either LasR or LasI even nullified the expression of genes regarding Iqs system, and thus, reduced the production of 2-(2-hydroxyphenyl)-thiazole-4-carbaldeghyde (IQS) [32]. Therefore, the observation of the transcriptional expression of QS regulatory functional genes can provide the primary determinant of the QSI hierarchy system.
In this study, transcriptional expression of LasR, RhlR and PqsR genes were all down-regulated to 27%, 39% and 18%, respectively, in the presence of 1 mM vanillin (Fig. 6). The down-regulation of transcriptional expression of QS regulatory genes was probably due to the structural similarity of vanillin with QS signal molecules. In the previous study, vanillin possessed the direct interaction with AHL receptor, which further interfered the binding of QS signal molecules with and the QS receptors [33]. Thus, the mitigated biofilm formation on the FO membrane surface by the addition of vanillin was probably due to the down-regulation of transcriptional expression of QS regulatory genes as described in Fig. 7 [32, 34, 35].
4. ConclusionsIn this study, the addition of vanillin as the quorum sensing inhibitor was investigated. The addition of 1 mM vanillin did not induce any bacteriostatic and bactericidal activity to P. aeruginosa, but significantly reduced the biomass of biofilm on the FO membrane surface. The bioassay showed that 1 mM vanillin was not sufficient to degrade the QS signal molecules such as N-Hexanoyl-DL-homoserine lactone nor did not alter the expression of genes related to the synthesis of QS signal molecules. However, the addition of vanillin effectively down-regulated QS receptor genes due to the structural similarity of vanillin with QS signal molecules. It is proposed that the interconnected and hierarchical QS system regulated by QS receptor genes in P. aeruginosa result the mitigation of biofilm formation on the surface of FO membrane. The results of this study implied the potential use of plant-oriented organic molecules as quorum sensing inhibitors in membrane processes where the concentration of organic matters might not be important.
AcknowledgmentThis work was supported by Korea Environment Industry & Technology Institute (KEITI) through Industrial Facilities & Infrastructure Research Program, funded by Korea Ministry of Environment (MOE) (1485016279). This work was also supported by the National Research Foundation of Korea Grant funded by the Ministry of Science, ICT and Future Planning (MSIP) (NRF-2017R1A2B3006354) through the National Research Foundation of Korea. This article was presented at the 2019 International Desalination Workshop (IDW2019) held on 28–30 August 2019, Jeju, Korea. And each individual author in this article contributed as below.
NotesAuthor Contributions S-J.C. (Ph.D. student) conducted all the experiments and wrote the manuscript. D-K.P. (M.S.) conducted the experiments and the data curation. D.J. (Post Doctor) set up the methodology of the experiments. Y-M.Y. (Assistant Professor) validated the results and revised the manuscript. S.K. (Associate Professor) supervised the entire experiments and revised the manuscript. References1. Shannon MA, Bohn PW, Elimelech M, Georgiadis JG, Marinas BJ, Mayes AM. Science and technology for water purification in the coming decades. Nature. 2008;452:301–310.
2. Ansari AJ, Hai FI, Price WE, Drewes JE, Nghiem LD. Forward osmosis as a platform for resource recovery from municipal wastewater - A critical assessment of the literature. J Membr Sci. 2017;529:195–206.
3. Herzberg M, Kang S, Elimelech M. Role of Extracellular Polymeric Substances (EPS) in Biofouling of Reverse Osmosis Membranes. Environ Sci Technol. 2009;43:4393–4398.
4. Lin H, Zhang M, Wang F, et al. A critical review of extracellular polymeric substances (EPSs) in membrane bioreactors: Characteristics, roles in membrane fouling and control strategies. J Membr Sci. 2014;460:110–125.
5. Yoon H, Baek Y, Yu J, Yoon J. Biofouling occurrence process and its control in the forward osmosis. Desalination. 2013;325:30–36.
6. Wang Z, Tang J, Zhu C, Dong Y, Wang Q, Wu Z. Chemical cleaning protocols for thin film composite (TFC) polyamide forward osmosis membranes used for municipal wastewater treatment. J Membr Sci. 2015;475:184–192.
7. Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science. 1998;280:295–298.
8. Yeon K-M, Cheong W-S, Oh H-S, et al. Quorum Sensing: A New Biofouling Control Paradigm in a Membrane Bioreactor for Advanced Wastewater Treatment. Environ Sci Technol. 2009;43:380–385.
9. Kim S-R, Oh H-S, Jo S-J, et al. Biofouling control with bead-entrapped quorum quenching bacteria in membrane bioreactors: physical and biological effects. Environ Sci Technol. 2013;47:836–842.
10. Lee SH, Lee S, Lee K, et al. More Efficient Media Design for Enhanced Biofouling Control in a Membrane Bioreactor: Quorum Quenching Bacteria Entrapping Hollow Cylinder. Environ Sci Technol. 2016;50:8596–8604.
11. Nahm CH, Choi D-C, Kwon H, et al. Application of quorum quenching bacteria entrapping sheets to enhance biofouling control in a membrane bioreactor with a hollow fiber module. J Membr Sci. 2017;526:264–271.
12. Yu H, Lee K, Zhang X, Choo K-H. Core-shell structured quorum quenching beads for more sustainable anti-biofouling in membrane bioreactors. Water Res. 2019;150:321–329.
13. Plyuta V, Zaitseva J, Lobakova E, Zagoskina N, Kuznetsov A, Khmel I. Effect of plant phenolic compounds on biofilm formation by Pseudomonas aeruginosa. APMIS. 2013;121:1073–1081.
14. Jia P, Xue YJ, Duan XJ, Shao SH. Effect of cinnamaldehyde on biofilm formation and sarA expression by methicillin-resistant Staphylococcus aureus. Lett Appl Microbiol. 2011;53:409–416.
15. Kumar L, Chhibber S, Harjai K. Zingerone inhibit biofilm formation and improve antibiofilm efficacy of ciprofloxacin against Pseudomonas aeruginosa PAO1. Fitoterapia. 2013;90:73–78.
16. Ponnusamy K, Paul D, Kweon JH. Inhibition of Quorum Sensing Mechanism and Aeromonas hydrophila Biofilm Formation by Vanillin. Environ Eng Sci. 2009;26:1359–1363.
17. Kappachery S, Paul D, Yoon J, Kweon JH. Vanillin, a potential agent to prevent biofouling of reverse osmosis membrane. Biofouling. 2010;26:667–672.
18. Choi S-J, Park D-K, Yun Y-M, Kang S. Inhibition of biofilm formation on FO membrane surface by plant-oriented organic molecules. Desalin Water Treat. 2017;99:112–116.
19. Sadr Ghayeni SB, Beatson PJ, Schneider RP, Fane AG. Adhesion of waste water bacteria to reverse osmosis membranes. J Membr Sci. 1998;138:29–42.
20. O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci. 2013;110:17981–17986.
21. Vandeputte OM, Kiendrebeogo M, Rajaonson S, et al. Identification of Catechin as One of the Flavonoids from Combretum albiflorum Bark Extract That Reduces the Production of Quorum-Sensing-Controlled Virulence Factors in Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2010;76:243–253.
22. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10:57.
23. Güell M, Yus E, Lluch-Senar M, Serrano L. Bacterial transcriptomics: what is beyond the RNA horiz-ome? Nat Reviews Microbiol. 2011;9:658.
24. Choi S-J, Jang D, Lee J-K, Yun Y-M, Kang S. Mitigation of biofouling in forward osmosis process by bacteria-oriented quorum quenching molecules. Desalin Water Treat. 2017;99:107–111.
25. Choo JH, Rukayadi Y, Hwang JK. Inhibition of bacterial quorum sensing by vanilla extract. Lett Appl Microbiol. 2006;42:637–641.
26. Chugani SA, Whiteley M, Lee KM, D’Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci. 2001;98:2752–2757.
27. Han S-S, Kim WJ, Hong Y, et al. RNA sequencing identifies novel markers of non-small cell lung cancer. Lung Cancer. 84:229–235.
28. Valenzuela-Miranda D, Del Río-Portilla MA, Gallardo-Escárate C. Characterization of the growth-related transcriptome in California red abalone (Haliotis rufescens) through RNA-Seq analysis. Mar Genomics. 2015;24:199–202.
29. Yemiş GP, Pagotto F, Bach S, Delaquis P. Effect of Vanillin, Ethyl Vanillin, and Vanillic Acid on the Growth and Heat Resistance of Cronobacter Species. J Food Prot. 2011;74:2062–2069.
30. National Center for Biotechnology Information (NCBI). Gene [Internet]. Bethesda (MD): National Library of Medicine (US); c2020. [cited 13 July 2020]. Available from: https://www.ncbi.nlm.nih.gov/gene/
31. Lu C, Kirsch B, Zimmer C, et al. Discovery of Antagonists of PqsR, a Key Player in 2-Alkyl-4-quinolone-Dependent Quorum Sensing in Pseudomonas aeruginosa. Chem Biol. 2012;19:381–390.
32. Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6:26–41.
33. Teplitski M, Robinson JB, Bauer WD. Plants Secrete Substances That Mimic Bacterial N-Acyl Homoserine Lactone Signal Activities and Affect Population Density-Dependent Behaviors in Associated Bacteria. Mol Plant-Microbe Interact. 2000;13:637–648.
Fig. 2Bacteriostatic (a) and bactericidal effect (b) of P. aeruginosa at various concentration of vanillin. 
Fig. 3The changes in biomass of virgin and fouled FO membrane in the presence and absence of 1 mM vanillin after 24 h and 48 h. 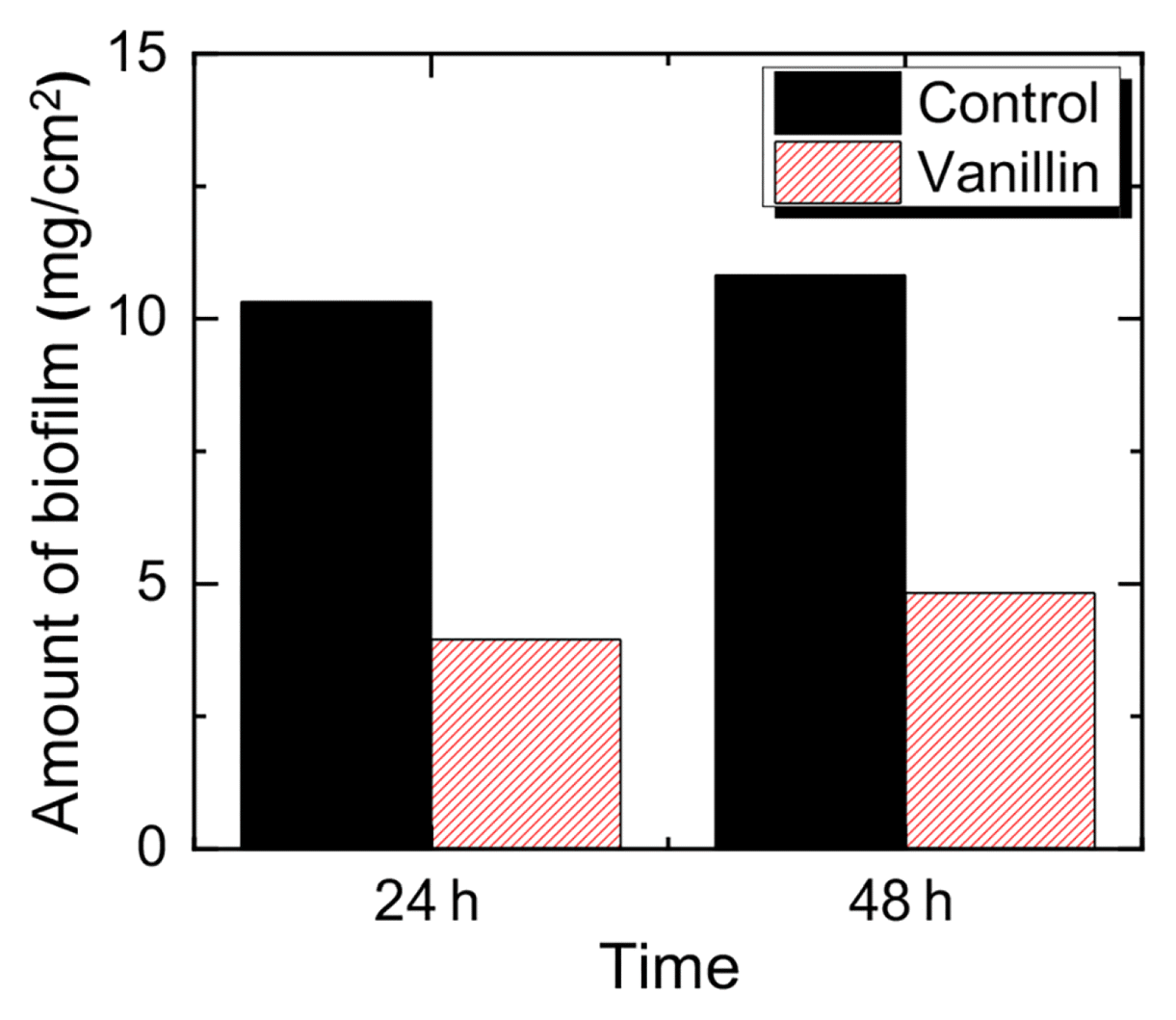
Fig. 5Expression of AHL-based QS genes (a) and quinone-based QS genes (b) in the presence and absence of 1 mM vanillin. The dashed line indicates the expression of genes equals to that of control. 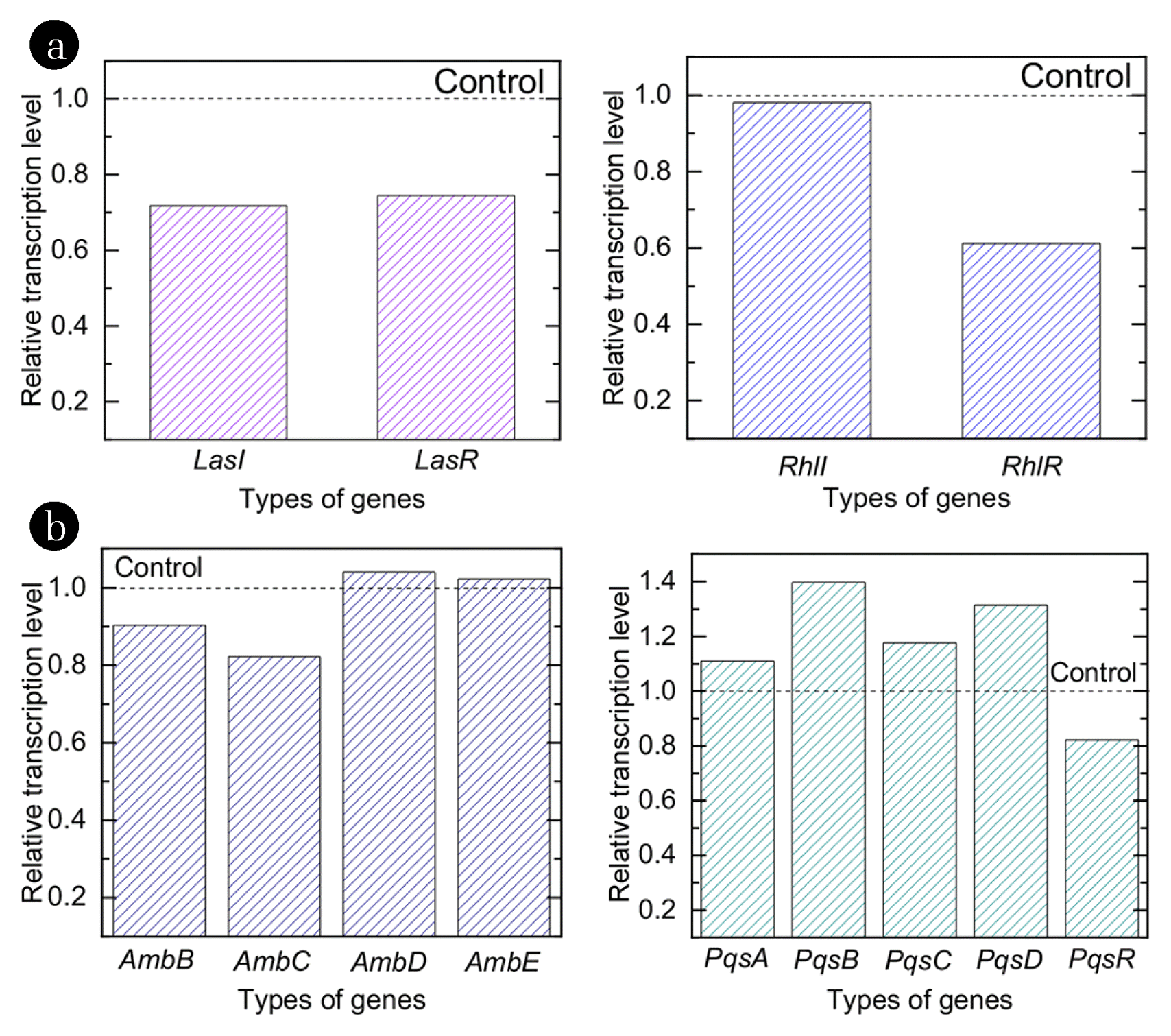
|
|
||||||||||||||||||||||||||||||||||||||