AbstractEffects of heavy metals on phenol-utilizing microorganisms that could co-metabolically degrade naphthol were investigated. Polymerase chain reaction (PCR) and denaturing gradient gel electrophoresis (DGGE) identified these phenol-utilizing microorganisms as Alicycliphilus denitrificans K601, Alicycliphilus sp. R-2461, uncultured Alicycliphilus sp., and Acidovorax aerodenitrificans. Inhibitory effects of Pb, Cu, Cd, and Zn on phenol biodegradation were in the order of Cu > Cd > Pb > Zn. Inhibitory effects of mixed heavy metals on phenol-utilizing microorganisms were in the order of (Cd + Pb) ≈ (Cd + Cu) > (Zn + Pb) > (Zn + Cd) > (Pb + Cu) > (Zn + Cu). The presence of mixed heavy metals synergistically inhibited microbial degradation of phenol. The relationship between reaction constants and inhibition concentrations that caused 50% of the degradation rate (IC50) for mixed heavy metals was derived as k2 = 0.69 · IC50−1.007. In this study, heavy metals inhibited phenol biodegradation through synergistic interactions. However, other studies have reported antagonistic interactions depending on the microbial community and heavy metals, indicating that the inhibition type of heavy metal on microbial degradation depends on the microorganism and the type of heavy metal.
1. IntroductionAlong with the increase in demand for various chemicals, energy, and agricultural and industrial products due to the process of many industries, such as refinery, pharmaceutical manufacturing industry, or chemical industry, many pollutants are spilling into the environment, resulting in impoverished living circumstances and increasing environmental pollution. Phenol, one of the pollutants generated in various processes, is an aromatic compound having a benzene ring structure. It accumulates in soil, rivers, and groundwater due to artificial contamination, causes toxicity to both animals and plants, and is not easily decomposed in nature, resulting in a social problem [1–4]. Petroleum refining, petrochemical industry, phenolic resin, pharmaceutical companies, coal conversion plants, and electronics industry plants are the main sources of pollution [3, 5]. Various physicochemical methods, such as chemical oxidation, solvent extraction, and adsorption by activated carbon, are applied to the removal of phenol from wastewater. However, the biological treatment of phenol using phenol-utilizing microorganisms is more economical and efficient than the physicochemical method and has no fear of secondary contamination [4, 5].
Phenol can be degraded by various strains under aerobic and anaerobic conditions. Phenol-degrading strains mainly found in soil or activated sludge processes include Rhodococcus, Mycobacterium, Flavobacterium, Rhodococcus chlorophenolicus, Pseudomonas putida, Pseudomonas, Pikettii, Cryptococcus elinovi, Phanerochaete chrysosporium, Bacillus stearothermophilus, Coprinus cinereus, Alcaligenes sp., Acinetobacter sp., and green alga Scenedesmus abundans [2, 3, 6, 7]. The decomposition of organic substances by microorganisms is achieved by oxidation-reduction reactions of enzymes present in the bacterial body, and these microorganisms are reported to be capable of organic decomposition with or without halogen [8]. Phenol-degrading microorganisms can oxidize phenolic compounds to catechol by phenol hydroxylases and phenol monooxygenases, and catechol can be oxidized to cis-muconate by catechol 1,2-dioxygenase, ortho-cleavage pathway, and catechol 2,3-dioxygenase. Decomposition proceeds through the meta-cleavage pathway to 2-hydroxymuconic aldehyde, which can be finally converted to carbon dioxide [9–12]. It is noteworthy that phenol-utilizing microorganisms not only can decompose phenolic compounds that adversely affect the environment but also can detoxify xenobiotic compounds that are artificially contaminated in nature using oxidative enzymes of these microorganisms. In particular, it is known that phenol-utilizing bacteria can be used for co-metabolism decomposition of non-degradable organic substances such as trichloroethylene (TCE), dichloroethylene (DCE), vinyl chloride (VC), and naphthalene [13, 14]. Naphthol is a typical compound that is widely used in the manufacture of dyes and pesticides. It is known as a harmful substance to humans. It is a persistent compound due to biodegradation problem. However, few studies have investigated cometabolic degradation of naphthol in the presence of phenol utilizing bacteria. Along with its high potential toxicity, mutagenicity, and carcinogenicity of naphthol, studies on comebolic degradation of naphthol such as chlorinated hydrocarbons and naphthalene need to be conducted and the capability of a comebolic transformation process of phenol-utilizing bacteria needs to be confirmed [15–19].
In the biological decomposition of organic compounds, many factors can affect the decomposition ability or metabolism of microorganisms by preventing or stimulating the growth of organisms. These factors include temperature, pH, oxygen content and the bioavailability of contaminants to microorganisms, substrate concentration, and physical properties [20–22]. In addition to the general factors that influence phenol degradation listed above, the presence of heavy metals as co-contaminants can severely affect the biological decomposition of phenol [23, 24]. Many studies have been conducted on the negative effects of heavy metals on biological degradation, and some inhibition can be reduced through bio-sorption, bio-accumulation, or solidification of heavy metal; however, the negative effect of heavy metals on enzymes can affect microbial activity [25–27]. Considering the possibility that heavy metals may be discharged together with phenol in wastewater [28, 29], it is important to investigate the biological decomposition of phenol, and the effects of heavy metals on biodegradation [30].
In this study, the microbial enrichment that was capable of decomposing phenol was attempted from sewage sludge, and the degradation characteristics were investigated. The induction process of phenolic microorganisms involved in the co-metabolism was confirmed using naphthol, and the effects of heavy metals as co-contaminants on the biodegradation of phenol were examined. The negative effects of the phenol degradation with heavy metals were separately investigated in the case of the presence of single or mixed heavy metals. The identification of phenol-utilizing bacteria was attempted through PCR amplification, cloning, and sequencing of 16s ribosomal deoxyribonucleic acid (rDNA). The results of this study can be provided as reference data that can be used in the field as a complex analysis of the effects of heavy metals on the degradation characteristics of phenol-utilizing microorganisms.
2. Experimental Methods2.1. Source of a Sludge Sample and Enrichment of Phenol-utilizing MicroorganismsA sample was collected from an activated sludge process of a sewage treatment plant at Uijeongbu, South Korea for the enrichment of phenol-utilizing bacteria. Enrichment solution contained (per liter) 0.50 g NH4NO3, 0.50 gKH2PO4, 0.50 g MgSO4·7H2O, 0.10 g CaCl2, 0.50 g K2HPO4, 0.20 g NaCl, 0.01 g MnSO4·7H4O, 0.01 g FeSO4·7H2O, 0.05 g Yeast Extract, and phenol [8]. Before the experiment, a glass container and serum bottle were sterilized in an autoclave, to prevent contamination by other microorganisms. The enrichment solution (100 mL) and 1 mL of a sample collected from the activated sludge were added into a serum bottle and incubated at 113 rpm and 32°C in a shaking incubator. During the incubation period of the microorganism, the initial pH was 7.3. Three successive subcultures were performed before the phenol degradation experiment.
2.2. Phenol Degradation in the Presence of Naphthol or Heavy MetalsFor the phenol degradation experiment, an enrichment solution containing about 700 mg/L of phenol was placed in a serum bottle under aerobic conditions. Subcultured microorganisms were inoculated into the serum bottle. The inoculation amount of the seeding microorganism was 1% (V/V) of the total volume. To keep the activity of inoculated microorganisms the same in each experiment, subcultured microorganisms during the exponential growth phase were inoculated into the serum bottle. Samples were collected at an appropriate time during the one-week incubation period and refrigerated before analysis. To verify the induction of oxidase in microorganisms, 50 mg/L of naphthol was cultured alone or with phenol. Degradation patterns of phenol and naphthol were then examined. A blank sample (without inoculation of microorganisms) was prepared to observe the possibility of abiotic degradation of phenol or naphthol. Cd, Pb, Zn, and Cu were used to observe effects of heavy metals on phenol degradation. The composition of complex heavy metals was (Zn + Cd), (Pb + Zn), (Zn + Cu), (Cd + Pb), (Cd + Cu), and (Cu + Pb). The concentration of the complex heavy metal was maintained at the same concentration as the heavy metal alone. In the degradation test of phenol and naphthol, samples were collected once a day. For phenol and heavy metal tests, samples were collected three or four times a day.
2.3. PCR/DGGE and Microbial Identification AnalysisSamples collected for microbial identification analysis were centrifuged at 11,000 rpm for 5 min, and the precipitates excluding the supernatant were extracted using the Power Soil DNA Isolation Kit (MO BIO Laboratories, Inc. Korea), following the instructions of the manufacturer [31]. For 16S rDNA amplification, a first PCR was performed using 10f and 1400r primers, while a second PCR was performed with 341f and 518r primers. In addition, to perform DGGE, the second PCR has performed again by attaching a guanine and cytosine (GC) clamp to the 341f [32]. DGGE was performed using a DCode universal mutation detection system (BIO-RAD, DCode System CA, USA). PCR products were mixed with a DNA loading buffer and loaded into an 8% polyacrylamide (denaturant gradient between 40% and 60%). DGGE was performed at 200 V for 5 h at 60°C. Nucleotide sequencing of 16S rDNA was performed by Bionics Co., Ltd(Republic of Korea). The National Center for Biotechnology Information Basic Local Alignment Search Tool (NCBI BLAST) was used for the identification of each species in the microbial community [33–36].
2.4. Chemical Analysis and Toxicity Index of Heavy MetalsSamples for each experiment were centrifuged at 12,000 rpm to remove solids, and the supernatant was purified using high-performance liquid chromatography (HPLC, Yonglin YL9100, Korea), in order to analyze phenol and naphthol. For HPLC analysis, a C18 column (Waters 5μm, USA) and a ultraviolet (UV) detector with a wavelength of 254 nm were used [8]. The mobile phase was mixed with methanol: water at (60:40). Heavy metal was analyzed by Atomic Absorption Spectrophotometry (AAS, Perkin Elmer, USA) [37]. A sample volume of 10 mL was collected and filtered with a syringe membrane filter (0.45 μm), and then 1 mL of about 60 % nitric acid was put in a polyethylene bottle and stored frozen. Analysis of the experimental results was done using the average values obtained from repeated experiments.
2.4.1. Effects of heavy metals on phenol degradationThe reaction rate constant was calculated by regression analysis of the phenol concentration using Eq. (1). IC50 was determined by the concentration of heavy metals that reached half of the initial rate constant (in the absence of heavy metal) [38]:
where, C = phenol concentration (mg/L), t = time (h), k = reaction constant (h−1).
2.4.2. Additive toxicity index of mixed heavy metals on phenol degradationTo evaluate the interaction between heavy metals, the additive toxicity index (AI) method was used. The additive toxicity index method is a method of evaluating the contribution of heavy metal mixtures after obtaining the IC50 value of each heavy metal and the IC50 value when heavy metals are mixed. The calculation formula of Eq. (2) was used [39]:
where, S = sum of toxic reactions, Am = IC50 of substance A from mixed heavy metal, Bm = IC50 of substance B from mixed heavy metal, Ai = IC50 of substance A from single heavy metal, Bi = IC50 of substance B from single heavy metal.
Using the S value calculated in the above equation, the correlation between the two heavy metals can be evaluated based on the AI value. When the S value is less than 1, the AI calculation is 1/S-1.0; when the S value is greater than 1, -S + 1.0; and when the S value is 1, it is calculated as S-1 (= 0). If the calculated AI value is greater than 0, it is a synergistic interaction between heavy metals; and if it is less than 0, it is an antagonistic interaction [39]. Synergistic interaction means the effect of two heavy metals taken together is greater than the sum of their separate effects at the same dose. Antagonistic interaction means the effect of two heavy metals taken together is less than the sum of their separate effects at the same dose. This is because the second heavy metal increases the excretion of the first, or even directly blocks its toxic actions.
3. Results and Discussions3.1. Characteristics of Phenol and Naphthol DegradationTo evaluate the phenol degradation of the microorganisms the phenol biodegradation over time in a medium composed of about 700 mg/L phenol as a carbon and energy source was analyzed. Fig. 1 shows that when the microorganism was not inoculated, the change in the concentration of phenol and naphthol with time was hardly observed, which means that under this experimental condition, the abiotic decomposition of phenol or naphthol does not occur. When microorganisms were inoculated, about 700 mg/L phenol was completely decomposed in 3 d in the culture medium containing only phenol as a carbon and energy source. In the case of inoculated samples of microorganisms that used only naphthol as a carbon and energy source, the change in naphthol concentration was similar to that of the blank sample. This slight change indicates that no biodegradation of naphthol occurred. When phenol and naphthol were present together, 676.16 mg/L of phenol was decomposed in 7 d, and 53.19 mg/L of naphthol was reduced to 19.58 mg/L in the sample, and about 33.61 mg/L of naphthol was decomposed. This indicates the co-metabolism of naphthol by phenolic microorganisms, indicating that naphthol alone is not degraded, but the enzyme induction by the oxidase produced by the degradation of phenol proceeds.
The decomposition of these co-metabolic non-degradable substances is the same type of co-metabolism that occurs in ammonia monooxygenase, toluene, or butanoxylase, and it is reported that the decomposition of TCE or naphthalene can proceed by co-metabolic process [13, 14, 40, 41]. However, it is noteworthy that Fig. 1 shows that the degradation rate of phenol was significantly slower in the mixed sample of phenol and naphthol than in the degradation rate of phenol when phenol alone was used. This is consistent with the results of the previous study that when the same enzyme simultaneously decomposes the secondary substrates, the degradation of the primary substrate proceeds slowly [8].
3.2. DGGE and Sequencing Analysis for the Identification of Phenol-utilizing MicroorganismAfter amplifying 16s-rDNA through PCR to identify the enrichment cultured phenol-utilizing microorganism, DGGE was performed. Table 1 summarizes sequence affiliation of each band on the DGGE profile (Fig. S1) based on NCBI BLAST analysis. All four types of bands presented as phylogenetically belonging to β-proteobacteria. The cluster analysis results showed that Bands 1, 2, 3, and 4 were Alicycliphilus denitrificans K601, Alicycliphilus sp. R-2461 hits, Uncultured Alicycliphilus sp., and Acidovorax aerodenitrificans hits, respectively, among which Alicycliphilus denitrificans K601 strains have been reported to degrade phenol, benzene, and toluene [10, 42]. Alicycliphilus denitrificans K601 is a short rod-shaped aerobic gram-negative bacterium that can grow on aerobic sewage sludge. The optimum pH range lies in the range 7.2–7.4, while the optimum temperature is known to range 30–37°C [43]. In the case of Bands 2 and 3, it is judged that there is ecological significance in the genus Alicycliphilus like Band 1, but in the case of Acidovorax aerodenitrificans in Band 4, no data on the microbial community have previously been reported. This is the first reported species on phenol degradation.
3.3. Effect of Heavy Metals on Phenol DegradationTo investigate the effect of heavy metals on the phenol biodegradation, heavy metals were added into the medium at a certain concentration, and then the degradation patterns of phenol were examined using a first-order reaction. After adding Cu, Cd, Pb, and Zn independently, microorganisms were inoculated, and the inhibitory effect of phenol degradation according to the single heavy metal was compared. Fig. 2 shows the degradation pattern of phenol under Zn of 0.05, 0.2, 2.6, 10, or 20 mg/L, Cd of 1, 5, or 20 mg/L, Pb of 1, 5, 20, 30, or 40 mg/L, or Cu of 0.5, 1, 2, 5, or 10 mg/L, with initial concentration of phenol at about 700 mg/L. In each experiment, it was found that the phenol degradation rate was inhibited, and as the concentration of the heavy metal increased, slowed down. The degradation rate constant (k) was obtained by the regression analysis using Eq. (1). It was found that the higher the heavy metal concentration, the lower the reaction constant value
The IC50 was calculated as the heavy metal concentration at which the degradation rate constant value (4.4 h−1) of phenol produced in the absence of heavy metals was inhibited by 50%. In order to calculate IC50, another regression was accomplished by the first-order reaction of k vs heavy metal concentration at Fig. 3. Table 2 summarizes the reaction constants (k2, L/mg/h2). Higher reaction constants (k2) obtained by the regression at Table 2 represent higher inhibition of the degradation of phenol. The IC50 values of Pb, Cu, Cd, and Zn were measured to be 6.86, 2.77, 3.61, and 13.15 mg/L, respectively. Considering that the lower the inhibition by heavy metals shows the higher the IC50 value, these values show that the inhibitory effect on the phenol degradation appears in the order Cu > Cd > Pb > Zn. Heavy metals have been reported to inhibit the specific metabolic activities of microorganisms, and Table 2 compares the IC50 values presented to the results of previous studies and also summarizes the effects of such heavy metals in this study [39, 44–47]. The IC50 values of phenolic microorganisms for single heavy metals were found to represent approximately middle values within the range of previously reported IC50 values, but the effect of Cd is somewhat lower than in the others. According to the previous studies of the effect of individual heavy metals on the polycyclic aromatic compounds (PAH) degradation, it has been reported that the inhibition of microbial activity by Cu and Cd is higher than that of Zn and Pb [26, 48]. Our experimental results are similar to those results.
3.4. The Effect of Mixed Heavy Metals on the Phenol DegradationSince heavy metal is more likely to exist as mixed heavy metals than a single heavy metal in a common environment, this study attempted to analyze the effect of mixed heavy metals on phenol degradation. The mixed heavy metal was defined as the case where the observed Cd, Cu, Pb, and Zn in the inhibition of a single heavy metal were mixed. The cases were divided into six categories as (Zn + Cd), (Pb + Zn), (Zn + Cu), (Cd + Pb), (Cd + Cu), and (Cu + Pb). Fig. 4 shows the degradation pattern of phenol according to the mixed heavy metals in each case and analyzed by the first-order reaction as Eq. (1). Similar to single heavy metals, the higher the concentrations of heavy metals, the slower the phenol degradation rate in the presence of mixed heavy metals. The concentrations under different cases were as follows:
Zn + Cd (0.05 + 1, 0.2 + 5, 2.6 + 20) mg/L; Pb + Zn (1 + 0.05, 5 + 0.2, 20 + 2.6, 30 + 10, 40 + 20); Zn + Cu (0.05 + 0.5, 0.2 + 1, 2.6 + 2, 10 + 5, 20 + 10) mg/L; Cd + Pb (1 + 1, 5 + 5, 20 + 20) mg/L; Cd + Cu (1 + 0.5, 5 + 1, 20 + 2) mg/L; and Cu + Pb (0.5 + 1, 1 + 5, 2 + 20, 5 + 30, 10 + 40) mg/L.
The degradation rate constant and IC50 of the mixed heavy metal were measured through regression analysis of the phenol degradation pattern. Fig. 5 confirms that the inhibitory effect on phenol degradation was in the order: (Cd + Pb) ≈ (Cd + Cu) > (Pb + Zn) > (Zn + Cd) > (Cu + Pb) > (Zn + Cu). Table 3 shows the IC50 value of the mixed heavy metal, resulting in the range (0.34–1.84), which was lower than the range of the IC50 value of the single heavy metal of range (2.77–13.15). This shows a synergistic interaction as a result of the interaction according to the mixed heavy metal present.
Table 3 also summarizes the AI values that were calculated using the IC50 values to confirm the interaction between single heavy metals. It was found that the reaction constant (k2) and the IC50 value obtained by the regression analysis of phenol degradation at Fig. 5 have an exponential correlation (reaction constant, k2 = 0.69 · IC50−1.007, R2 = 1). The AI value in Table 3 calculated according to the IC50 value was found to show synergistic interactions with mixed heavy metals. This means that when heavy metals are mixed, they have a greater inhibitory effect on the degradation of microorganisms, and in the presence of mixed heavy metals, the degradation rate of phenol has proceeded slower. Considering that the higher the inhibitory effect by the mixing of single heavy metals shows the higher AI value, these values show that the synergistic interaction by mixing of single heavy metal is severe in the order (Zn + Pb) > (Cd + Pb) > (Cd + Cu) > (Zn + Cd) ≈ (Zn + Cu) > (Pb + Cu) on phenol degradation.
The presence of mixed heavy metals has been reported to exhibit antagonistic inhibitory activity in other microorganisms, unlike in the case of phenol microbes that exhibit synergistic interactions in the phenol degradation [45, 49]. The variation of the IC50 values according to the microbial community indicates the effect could be very different depending on the microbe or type of heavy metal.
Therefore, when predicting the inhibition type of heavy metal on a certain microorganism, the type of microbial community or heavy metal should be considered in advance.
4. ConclusionsIn this study, the degradation pattern of phenol was investigated using a mixed strain that was capable of the degradation of phenol and naphthol by co-metabolism. The interactions of single or mixed heavy metals, such as Zn, Cd, Pb, and Cu, were evaluated with phenol degradation. The results obtained through this experiment are summarized below:
Naphthol was cometabolically degraded in the presence of phenol. When approximately 20 mg/L phenol was supplied, 1 mg/L naphthol could be degraded. Cluster analysis of phenol-utilizing microorganisms revealed Alicycliphilus denitrificans K601, Alicycliphilus sp. R-2461, uncultured Alicycliphilus sp., and Acidovorax aerodenitrificans belonging to β-proteobacteria.
IC50 values of Pb, Cu, Cd, and Zn were measured to be 6.86, 2.77, 3.61, and 13.15 mg/L, respectively. Inhibitory effects of heavy metals on phenol degradation were in the order Cu > Cd > Pb > Zn. Based on IC50 of mixed heavy metals for phenol-utilizing microorganisms, inhibitory effects were in the order (Cd + Pb) ≈ (Cd + Cu) > (Zn + Pb) > (Zn + Cd) > (Pb + Cu) > (Zn + Cu).
There was an exponential correlation between the reaction constant (k2) and the IC50 value obtained by regression analysis (reaction constant, k2 = 0.69·IC50−1.007, R2 = 1). Based on AI values calculated according to IC50 values, it was confirmed that all mixed heavy metals exhibited synergistic interactions.
AcknowledgmentsThis research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A0304274211).
NotesAuthor Contributions B.N (M.Sc. student) conducted all the experiments, analyzed the data, and wrote the manuscript. M.B (M.Sc. student) contributed to the design of the experiment and implementation of the research. N.Z.LM. (M.Sc. student) contributed to analyze data and revise the manuscript. T.J.L. (Professor) led the overall research as a corresponding author, supervised the student, and revised the manuscript. References1. Watanabe K, Teramoto M, Futamata H, Harayama S. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl Environ Microbiol. 1998;64:4396–4402.
2. Lee HD, Lee ME, Kim HG, Suh HH. Isolation of a phenol-degrading bacterial strain and biological treatment of wastewater containing phenols. J Life Sci. 2013;23:1273–1279.
3. Nair CI, Jayachandran K, Shashidhar S. Biodegradation of phenol. Afr J Biotechnol. 2008;7:4951–4958.
4. Lallement A, Besaury L, Tixier E, et al. Potential for phenol biodegradation in cloud waters. Biogeosci. 2018;15:5733–5744.
5. Hinteregger C, Leitner R, Loidl M, Ferschl A, Streichsbier F. Degradation of phenol and phenolic compounds by Pseudomonas putida EKII. Appl Microbiol Biotechnol. 1992;37:252–259.
6. Díaz E, Prieto MA. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr Opin Biotechnol. 2000;11:467–475.
7. Fawzy MA, Alharthi S. Cellular responses and phenol bioremoval by green alga Scenedesmus abundans: Equilibrium, kinetic and thermodynamic studies. Environ Technol Innov. 2021;22:101463.
8. Lee TJ, Kim SS, Chang SW. Cometabolic Biotransformation of Polyaromatic Hydrocarbon(PAH) using Phenol Acclimated Bacteria. J Korean Soc Environ Eng. 1997;19:1369–1378.
9. Mishra VK, Kumar N. Microbial degradation of phenol: a review. J Water Pollut Purif Res. 2017;4:17–22.
10. Oosterkamp MJ, Veuskens T, Plugge CM, et al. Genome sequences of Alicycliphilus denitrificans strains BC and K601T. J Bacteriol. 2011;193:5028–5029.
11. Zhang W, Yin K, Chen L. Bacteria-mediated bisphenol A degradation. Appl Microbiol Biotechnol. 2013;97:5681–5689.
12. Duraisamy P, Sekar J, Arunkumar AD, Ramalingam PV. Kinetics of phenol biodegradation by heavy metal tolerant rhizobacteria Glutamicibacter nicotianae MSSRFPD35 from distillery effluent contaminated soils. Front Microbiol. 2020;11:1573.
13. Suttinun O, Luepromchai E, Müller R. Cometabolism of trichloroethylene: concepts, limitations and available strategies for sustained biodegradation. Rev Environ Sci Biotechnol. 2013;12:99–114.
14. Abo-State MAM, Riad BY, Bakr AA, Aziz MA. Biodegradation of naphthalene by Bordetella avium isolated from petroleum refinery wastewater in Egypt and its pathway. J Radiat Res Appl Sci. 2018;11:1–9.
15. Liu XQ, Ding HS, Wang YY, Liu WJ, Jiang H. Pyrolytic temperature dependent and ash catalyzed formation of sludge char with ultra-high adsorption to 1-naphthol. Environ Sci Technol. 2016;50:2602–2609.
16. Mahugo Santana C, Sosa Ferrera Z, Esther Torres Padrón M, Juan Santana Rodríguez J. Methodologies for the extraction of phenolic compounds from environmental samples: new approaches. Molecules. 2009;14:298–320.
17. Waigi MG, Kang F, Goikavi C, Ling W, Gao Y. Phenanthrene biodegradation by sphingomonads and its application in the contaminated soils and sediments: a review. Int Biodeterior Biodegradation. 2015;104:333–349.
18. Ahire KC, Kapadnis BP, Kulkarni GJ, Shouche YS, Deopurkar RL. Biodegradation of tributyl phosphate by novel bacteria isolated from enrichment cultures. Biodegradation. 2012;23:165–176.
19. Zhou Y, Tang L, Yang G, et al. Phosphorus-doped ordered mesoporous carbons embedded with Pd/Fe bimetal nanoparticles for the dechlorination of 2, 4-dichlorophenol. Catal Sci Technol. 2016;6:1930–1939.
20. El-Naas MH, Al-Muhtaseb S, Makhlouf S. Biodegradation of phenol by Pseudomonas putida immobilized in polyvinyl alcohol (PVA), gel. J Hazard Mater. 2009;164:720–725.
21. Agarry SE, Durojaiye AO, Solomon BO. Microbial degradation of phenols: A review. Int J Environ Pollut. 2008;32:12–28.
22. Trigo A, Valencia A, Cases I. Systemic approaches to biodegradation. FEMS Microbiol Rev. 2008;33:98–108.
23. Duraisamy P, Sekar J, Arunkumar AD, Ramalingam PV. Kinetics of phenol biodegradation by heavy metal tolerant rhizobacteria Glutamicibacter nicotianae MSSRFPD35 from distillery effluent contaminated soils. Front Microbiol. 2020;11:1573.
24. Sokhn J, De Leij FAAM, Hart TD, Lynch JM. Effect of copper on the degradation of phenanthrene by soil micro-organisms. Lett Appl Microbiol. 2001;33:164–168.
25. Pratush A, Kumar A, Hu Z. Adverse effect of heavy metals (As, Pb, Hg, and Cr) on health and their bioremediation strategies: a review. Int Biotechnol. 2018;21:97–106.
26. Amor L, Kennes C, Veiga MC. Kinetics of inhibition in the biodegradation of monoaromatic hydrocarbons in presence of heavy metals. Bioresour Technol. 2001;78:181–185.
27. Yin K, Wang Q, Lv M, Chen L. Microorganism remediation strategies towards heavy metals. Chem Eng J. 2019;360:1553–1563.
28. Thasneema KK, Dipin T, Thayyil MS, et al. Removal of toxic heavy metals, phenolic compounds and textile dyes from industrial waste water using phosphonium based ionic liquids. J Mol Liq. 2021;323:114645.
29. Eletta OAA, Tijani IO, Ighalo JO. Adsorption of Pb (II) and phenol from wastewater using silver nitrate modified activated carbon from groundnut (Arachis hypogaea L.) shells. West Indian J Eng. 2020;43:26–35.
30. Pereira PP, Fernandez M, Cimadoro J, et al. Biohybrid membranes for effective bacterial vehiculation and simultaneous removal of hexavalent chromium (CrVI) and phenol. Appl Microbiol Biotechnol. 2021;105:827–838.
31. Vanparys B, Heylen K, Lebbe L, De Vos P. Pedobacter caeni sp. nov., a novel species isolated from a nitrifying inoculum. Int J Syst Evol Microbial. 2005;55:1315–1318.
32. Gao Y, Wang G, Zhou A, et al. Effect of nitrate on indole degradation characteristics and methanogenesis under mixed denitrification and methanogenesis culture. Biochem Eng J. 2019;145:33–41.
33. Auch AF, Henz SR, Holland BR, Goker M. Genome BLAST distance phylogenies inferred from whole plastid and whole mitochondrion genome sequences. BMC Bioinform. 2006;7:1–16.
34. Felgenstein J. Confidence limits on phylogenetics: an approach using the bootstrap. Evolution. 1985;39:783–791.
35. Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120.
36. Saitou N, Nei M. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425.
37. US Environmental Protection Agency. Hazardous Waste Test Methods/SW-846[Internet. February 2007. Available from: https://www.epa.gov/hw-sw846/sw-846-test-method-7000b-flame-atomic-absorption-spectrophotometry
39. Kim DH, Park SW, Kim DW, et al. Evaluation for Interactive Toxic Effects of Binary Heavy Metals on Bacterial Growth and Phosphorus Removal under Co-Culture Condition of Alcaligenes sp. and Pseudomonas sp. Appl Chem Eng. 2020;31:612–623.
40. Yang CE, Wu CY, Liu YC, Lan EI, Tsai SL. Cometabolic degradation of toluene and TCE contaminated wastewater in a bench-scale sequencing batch reactor inoculated with immobilized Pseudomonas putida F1. J Taiwan Inst Chem Eng. 2019;104:168–176.
41. Rolston HM, Hyman MR, Semprini L. Aerobic cometabolism of 1, 4-dioxane by isobutane-utilizing microorganisms including Rhodococcus rhodochrous strain 21198 in aquifer microcosms: Experimental and modeling study. Sci Total Environ. 2019;694:133688.
42. Solís-González CJ, Loza-Tavera H. Alicycliphilus: current knowledge and potential for bioremediation of xenobiotics. J Appl Microbiol. 2019;126:1643–1656.
43. Mechichi T, Stackebrandt E, Fuchs G. Alicycliphilus denitrificans gen. nov, sp. nov, a cyclohexanol-degrading, nitrate-reducing b-proteobacterium. Int J Syst Evol Microbiol. 2003;53:147–152.
44. Aidil MS, Sabullah MK, Halmi MI, et al. Assay for heavy metals using an inhibitive assay based on the acetylcholinesterase from Pangasius hypophthalmus (Sauvage, 1878). Fresenius Environ Bull. 2013;22:3572–3576.
45. Kim DH, Yoo J, Chung KY. Toxic Effects of Binary Mixtures of Heavy Metals on the Growth and P Removal Efficiencies of Alcaligenes sp. Korean J Environ Agric. 2016;35:79–86.
46. Janicki T, Długoński J, Krupiński M. Detoxification and simultaneous removal of phenolic xenobiotics and heavy metals with endocrine-disrupting activity by the non-ligninolytic fungus Umbelopsis isabellina
. J Hazard Mater. 2018;360:661–669.
47. Kim JH, Kim MG, Om AS. Verification of AGB (alpha-glucosidase biosynthesis) Bacterial Bioassay of Metal Toxicity and Inhibition Effect of Fulvic Acid (FA. ) Environ Health Toxicol. 2004;19:25–32.
Fig. 2Degradation of phenol in the presence of various concentrations of (a) Zn, (b) Cd, (c) Pb, or (d) Cu. 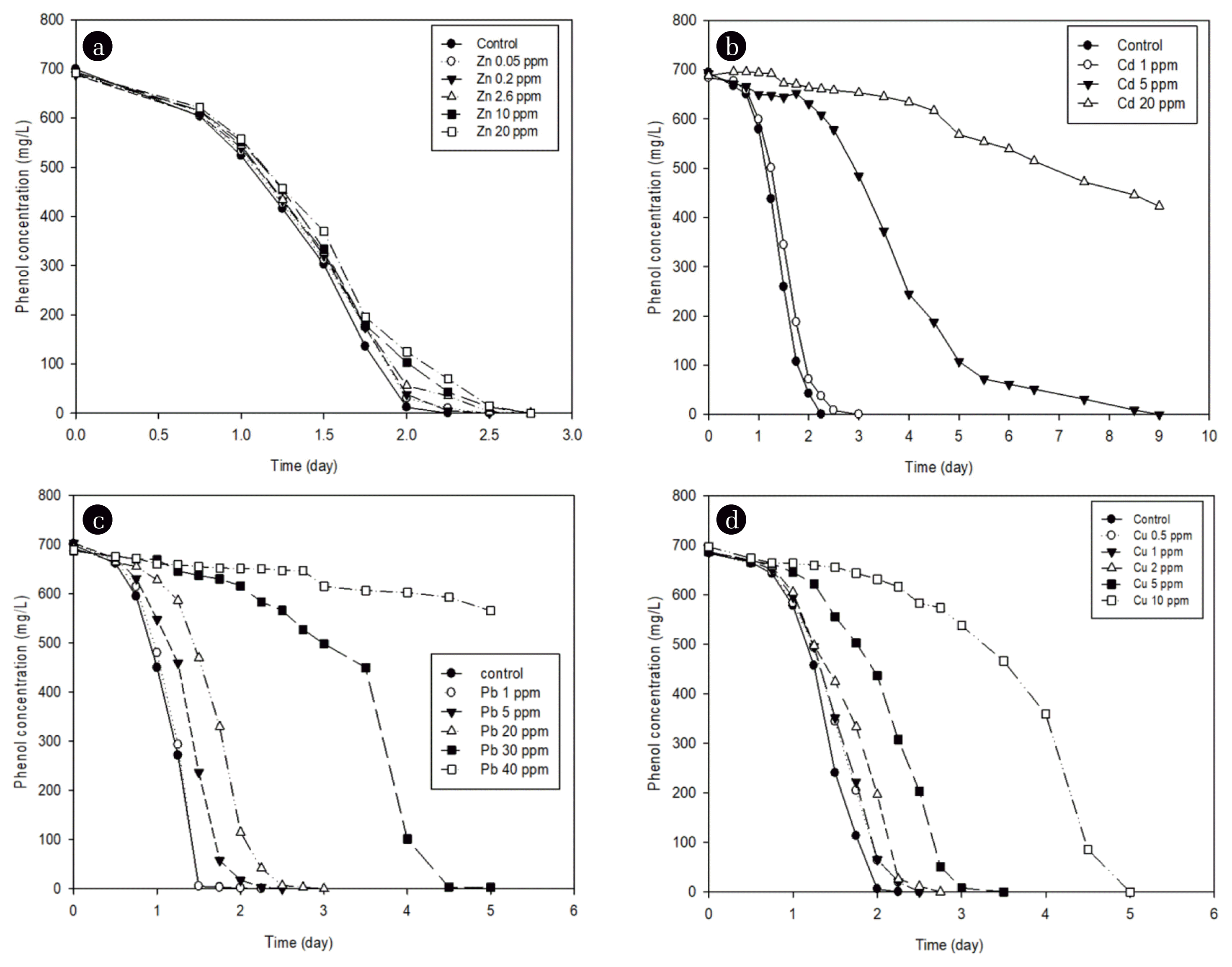
Fig. 3Variations of the phenol degradation rate and IC50 in the presence of Pb, Zn, Cu, and Cd. Black arrows on the graph represent the IC50 value of each heavy metal (a) Cu; (b) Cd; (c) Pb; (d) Zn. Regression was accomplished by the first-order reaction of k vs heavy metal concentration using Sigma plot ver. 12.5, Systat Inc., USA. 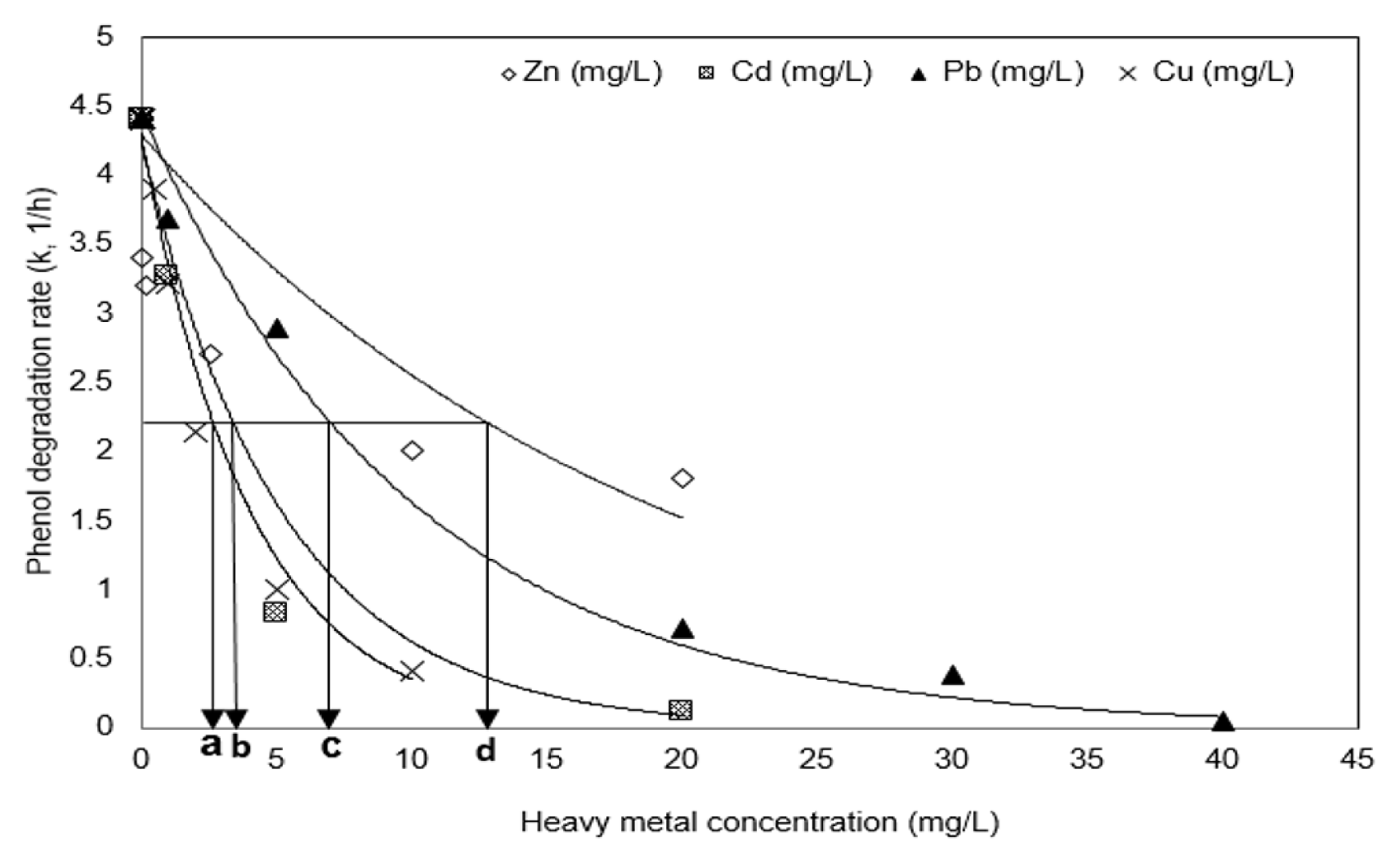
Fig. 4Degradation of phenol in the presence of various concentrations of mixed heavy metals (a) Zn + Cd; (b) Pb + Zn; (c) Zn + Cu; (d) Cd + Pb; (e) Cd + Cu; (f) Cu + Pb. 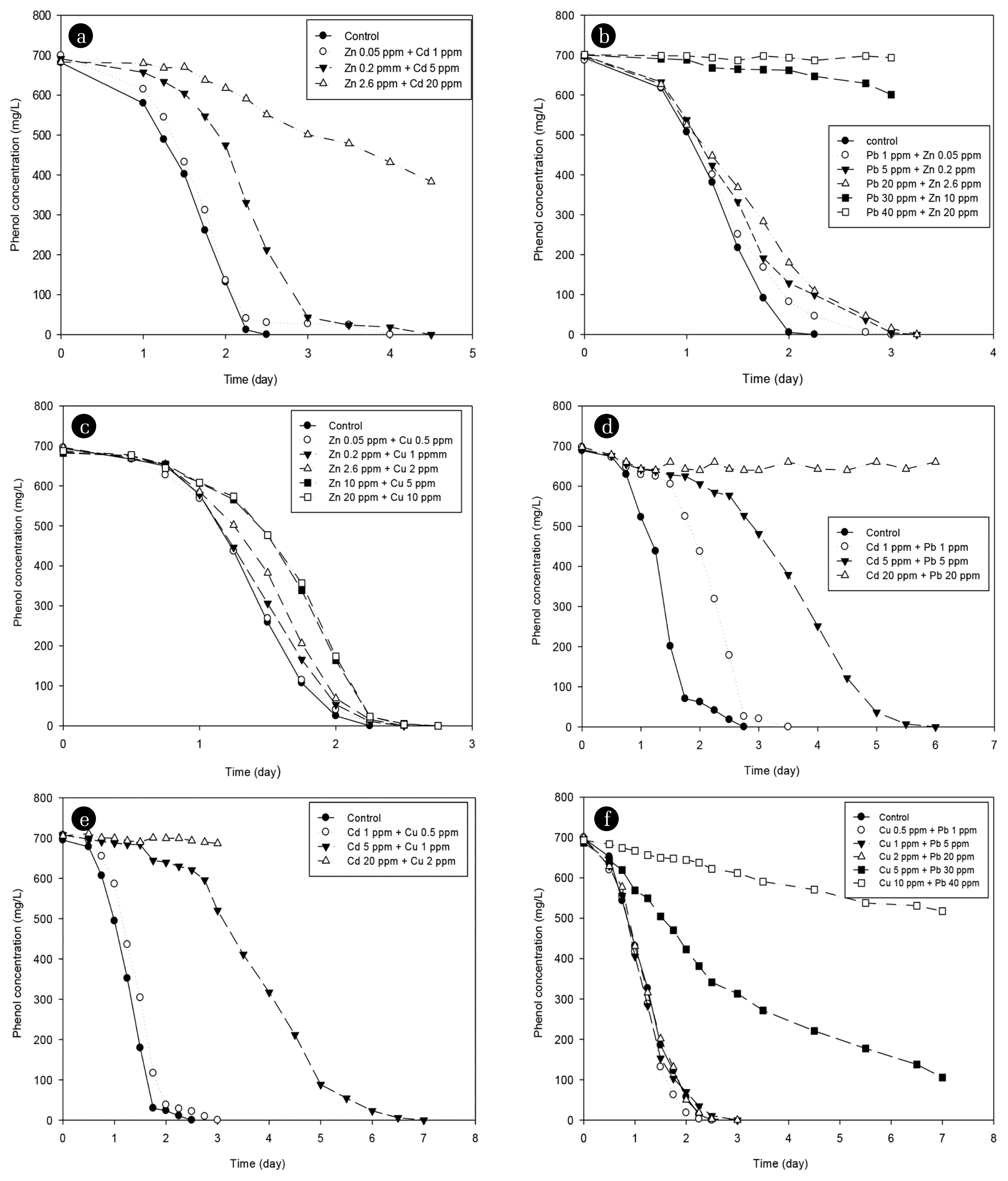
Fig. 5Variations of phenol degradation constants in the presence of the heavy metals and IC50 of the mixed heavy metals (a) Cd + Pb; (b) Cd + Cu; (c) Pb + Zn; (d) Zn + Cd; (e) Cu + Pb; (f) Zn + Cu. Regression was accomplished by the first-order reaction of k vs. mixed heavy metal concentration. 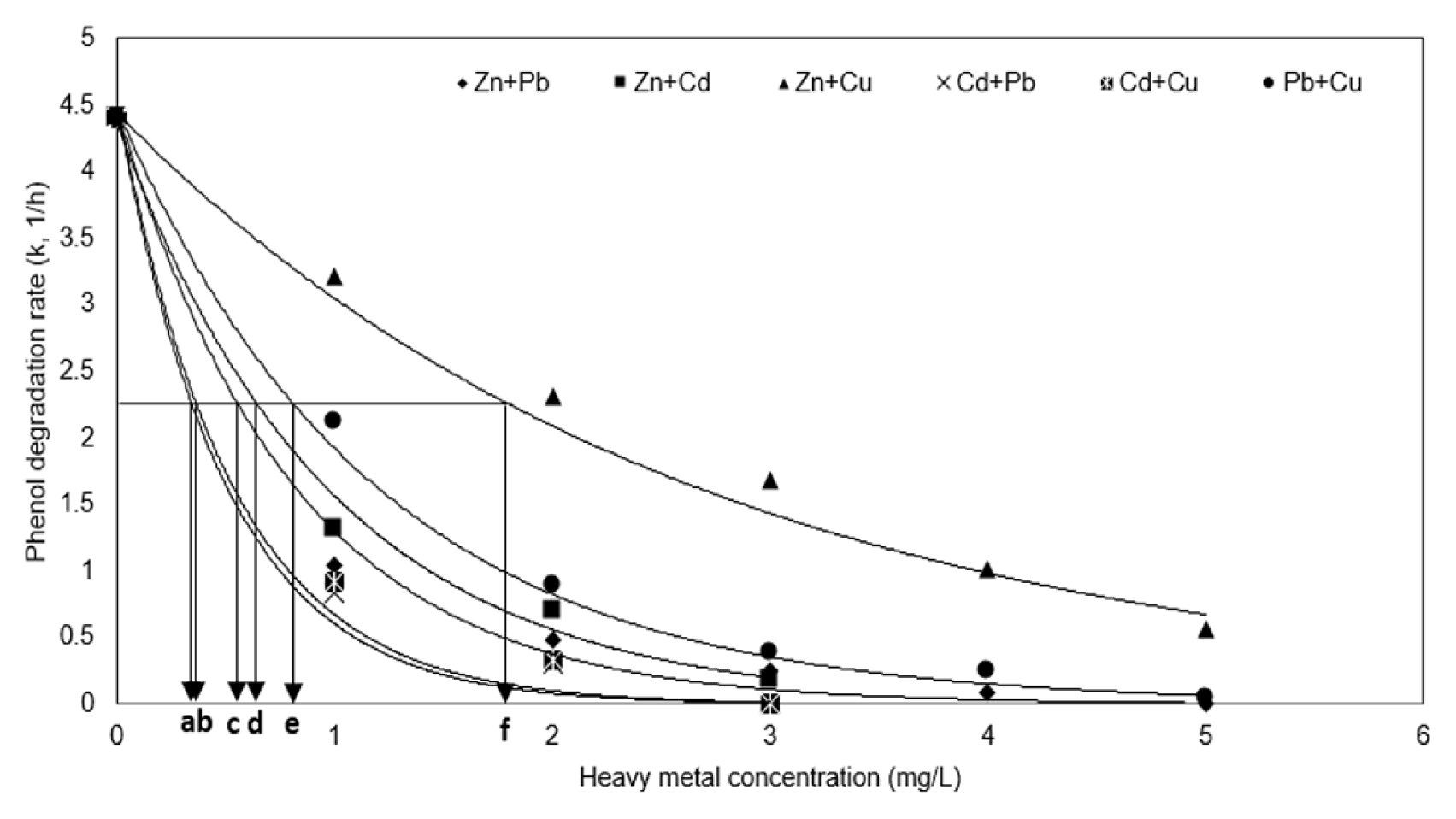
Table 1Affiliation of Denaturing Gradient Gel Electrophoresis (DGGE) Fragments Determined by Their 16S rDNA Sequence Table 2Comparison of IC50 Values with Those of Different Microorganisms
Table 3Summary of the Median Inhibitory Concentrations of Heavy Metals
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||