1. Introduction
Fast advancement, the growth of inhabitants, along with socio-economic progress have enormously increased the anthropogenic impact on the inherent ecosystems which are mainly responsible for environmental pollution [1]. 4-chlorophenol (4-CP) is listed among precedence contaminant by the US Environmental Protection Agency, owing towards carcinogenic and recalcitrant features [2]. 4-CP is widely used in various industries, including pulp and paper, leather tanning, biocide, dye, and herbicide industries as well as in chlorination of intake and waste water [3]. The effluent concentration of 4-CP in several manufacturing units differs within 100 to 1,000 mg/L [4]. 4-CP pollution severely affects the aquatic environment and may cause health-related problems such as diarrhea, amebiasis, dermal damage, respiratory problems, and several types of cancers [5]. Consequently, the anticipation of contamination of water resources along with shielding of communal health via conservation of water resources alongside the extent of ailments is an essential objective of wastewater treatment [6, 7].
The toxicity of 4-CP enforces the importance of 4-CP removal from industrial effluents. Several techniques have been investigated intended for the management of phenolic wastewater like adsorption, chemical oxidation, photo-degradation and solvent extraction, and advanced oxidation [8, 9]. However, the accumulation of toxic intermediate products along with low elimination competence of 4-CP are outstanding bottleneck issues of the existing traditional treatment strategies. On the contrary, biological treatment is advantageous than conventional treatment processes [10], owing to its high removal efficiency and the absence of the formation of toxic intermediates [11]. Therefore, inexpensive, eco-friendly biodegradation processes are desirable [12]. The formation of chloride ions (Cl−) during chlorophenol biodegradation aggravates toxicity to bacterial cells along with boosts the resistance towards the cleavage of aromatic rings [13].
Several species of Bacillus and Pseudomonas are reported to biodegrade 4-CP; however, most species can break down 4-CP at low concentrations. Therefore, the screening of the potential microbial strain(s) is an essential task for the efficient biodegradation of 4-CP. As the degradation potential of a biocatalyst may be assessed dependent on the kinetic parameters, knowledge of expansion along with biodegradation kinetics is essential meant for the forecasting of the sewage class and optimization of the reactor-functioning criteria to convene the release standards [14]. Most of the research studies have been focused on the isolation, screening, and characterization of microorganisms for the biodegradation of 4-CP. A few studies have been performed using isolated bacterial strains for biodegradation of 1,000 mg/L of 4-CP. However, very few studies have been directed to understand the kinetic nature of 4-CP biodegradation process. As per our knowledge concern, isolated bacterial strain used in this study, is the first bacterial species which can able to metabolize both 4-CP and phenol at the concentration of 1,000 mg/L as well as five different aromatic compounds. This metabolic adaptability creates the isolated strain an effective microorganism for the bioremediation of industrial effluents contaminated with diverse types of phenolic compounds. Hence, the potential significance of these parameters has been highlighted in the manuscript by experiments with isolated bacterial strain.
Here in this study, a bacterial strain was isolated from drainage water from a Hyundai car service center, Agartala, Tripura, India, and subjected to genetic characterization. The above strain was capable of rising on 4-CP along with utilization the mentioned substrate at an elevated concentration of 1,000 mg/L. The bacterial development and biodegradation kinetics of 4-CP were assessed with various kinetic parameters using different mathematical models.
2. Materials and Methods
2.1. Materials
4-CP (Sigma, USA), sodium chloride (Himedia, India), potassium nitrate (Himedia, India), nutrient broth (Himedia, India), magnesium sulfate (Sigma, USA), and agar powder (Himedia, India) were used in the current investigation. Every reagent utilized was an analytical standard and commercially accessible in India. GraphPad Prism 5 software was applied for nonlinear regression analysis to evaluate the model parameter used for kinetic analysis.
2.2. Culture Medium for Isolation Study
Isolation study was carried out using an inorganic medium supplemented with trace element solution. The media was composed of 0.5 g/L of ammonium nitrate (NH4NO3), 0.2 g/L of magnesium sulfate (MgSO4·7H2O), 0.5 g/L of dipotassium phosphate (K2HPO4), 0.5 g/L of monopotassium phosphate (KH2PO4), and 0.02 g/L of calcium chloride (CaCl2·2H2O). The trace element solution was supplemented to the inorganic medium at 10 mL/L and contained 0.3 g/L of ferrous sulfate (FeSO4·7H2O), 0.05 g/L of manganese sulfate (MnSO4·H2O), 0.1 g/L of cobalt chloride (CoCl2·6H2O), 0.034 g/L of sodium molybdate (Na2MoO4·2H2O), 0.04 g/L of zinc sulfate (ZnSO4), and 0.05 g/L of copper sulfate (CuSO4·5H2O) [15]. 4-CP was used at required concentration as the single source of carbon and energy. The bacterial strain was routinely transferred after ten days interval to an inorganic medium supplemented with 4-CP maintained as the single source of energy. pH of 7.4 was initially maintained during the preparation of media and incubation was performed at 37°C for 48 h. The working quantity of the medium was 50 mL for all experiments conducted in 250-mL Erlenmeyer flasks.
2.3. Isolation and Screening of 4-CP-degrading Strain
The bacterial strain used for 4-CP degradation was isolated from the drain outside the Hyundai denting and painting car service center, Agartala, Tripura, India. Ten milliliters of the sample was added to 50 mL of inorganic medium additionally amid 200 mg/L of 4-CP and kept at 37°C for 48 h in an incubator shaker at rpm of 120. 5 mL of previously incubated culture was transferred to new inorganic media via the similar developmental ambiance at every movement, apart from the 4-CP concentration elevation stepwise up to 1,000 mg/L and increment was carried out with 100 mg/L in each step. In each transfer, the absorbance of the culture medium (OD600) was observed at 600 nm from time to time. Acclimatization at each concentration of 4-CP was carried out three times. After acclimatization, the culture was incubated solid inorganic medium containing 1,000 mg/L of 4-CP. The streak plate method was applied to achieve the purified colonies. The purified colonies were moreover sorted intended for most excellent 4-CP degradation competency and used for further study.
2.4. 16S rDNA Gene Sequencing, Phylogenetic Analysis, and Thermodynamic Properties Analysis
Genomic analysis of the isolated strain was performed applying 16S rDNA technique. DNA was collected from the isolated species, and the quality of DNA was assessed on 1.2% agarose gel. The isolated DNA was amplified with 16S rRNA-specific primer using a thermal cycler. The PCR amplicon was enzymatically purified and further subjected to Sanger sequencing. Bi-directional DNA sequencing reaction of PCR amplicon was carried out with 8F and 1492R primers using BDT v3.1 cycle sequencing kit on ABI 3730xl Genetic Analyzer. The consensus sequence of 16S rDNA was deposited to the NCBI GenBank catalog, and a resemblance exploration was performed via the online BLAST platform (www.ncbi.nlm.nih.gov). Depends on the highest uniqueness value, first fifteen sequences were chosen and aligned applying ClustalW which is considered as multiple alignment software packages. The distance matrix was prepared via the Ribosomal record along with the highest likelihood phylogenetic tree was created. MEGA 5 was used to find out the evolutionary distance bootstrap data by the Jukes-cantor model of the neighbor-joining technique [16]. The thermodynamic characteristics of the bacterial strain were computed by an online package, (https://cail.cn/biotool/oligo/index.html).
2.5. Metabolic Adaptability Analysis
The metabolic adaptability of the species was estimated by inoculating the strain into inorganic medium additionally with various phenolic compounds such as phenol, 2-chlorophenol, 4-CP, 2,4-dichlorophenol, 2,4,6-trichlorophenol, 4-nitrophenol, and pentachlorophenol. The phenolic compounds were supplemented into the inorganic medium at a concentration of 100–1,000 mg/L. 10 mL of cells (OD600 ≈ 0.1) previously acclimatized at 1,000 mg/L of 4-CP was added at each flask and kept at a temperature of 37°C for 48 h in an incubator at rpm of 120. The residual concentrations of phenolic compound were estimated after 48 h of incubation by spectrophotometric method [11, 17].
2.6. Batch Kinetics of 4-CP Degradation
To evaluate kinetic factors, batch shake flask experiment was performed by adding ten milliliters of cells (OD600 ≈ 0.1) previously acclimatized at 1,000 mg/L of 4-CP into 50 mL of optimized inorganic medium containing trace element solution and kept at 37°C for 48 h in an incubator at rpm of 150. The composition of media used for biodegradation study was as follows: 0.75 g/L of ammonium nitrate (NH4NO3), 0.25 g/L of magnesium sulfate (MgSO4·7H2O), 0.75 g/L of dipotassium phosphate (K2HPO4), 0.25 g/L of monopotassium phosphate (KH2PO4), and 0.02 g/L of calcium chloride (CaCl2·2H2O). The trace element solution was supplemented as used for seed culture. 4-CP at the concentration of 1 g/L was used in the medium as the single source of energy along with carbon. Sampling was performed at 4 h interval for measurement of residual 4-CP and cell concentration. It is presumed that aeration is sufficient to provide the required oxygen levels and does not limit growth. Also, it was assumed that the growth of isolated bacteria is dependent on the concentration of 4-CP in the media at specified preliminary pH, temperature, along with the rate of aeration [18].
2.7. Analytical Procedures
UV-visible spectrophotometer was used for measurement of cell concentration by taking the absorbance at 600 nm wavelength. The optical densities (OD600) of the broth culture were converted as dry cell weight by plotting a calibration curve amid the dry cell weights versus optical density (OD600). The broth was centrifuged at 10,000 g for a predetermined duration of 10 min to separate cell biomass, and the sediment was washed and re-suspended [19]. It was further filtered via a pre-washed 0.45-mm filter paper and dried at 105°C until steady mass was found. For measurement of residual 4-CP, the broth was centrifuged for 10 min at 10,000 g, and 0.22-μm filter was used for filtration of the supernatant. The absorbance of the filtrate was measured at 298 nm wavelength using UV-visible spectrophotometer intended for the assessment of the enduring 4-CP concentration [11, 20].
3. Results and Discussion
3.1. Isolation and Characterization of 4-CP-degrading Strain
A 4-CP-degrading microbial strain was isolated from wastewater effluent of a Hyundai car service industry, Agartala, India. As the organic matter present in the effluent influence the concentration and variety of microorganisms in waste water, therefore, the organic-matter-rich waste water was chosen in the present study. Also, the waste water effluent from car service was thought to be rich in phenolic compounds; therefore, the isolation of 4-CP-degrading microorganism was carried out from the wastewater effluent of Hyundai car service industry, Agartala, India. Twenty-five effluent samples were collected from fifteen different places for the isolation of 4-CP-degrading bacteria. 10 mL of waste water was mixed to 50 mL of inorganic medium additionally amid trace element solution. Briefly, 200 mg/L of 4-CP was added into media, and the concentration of 4-CP was gradually increased. After two days of inoculation, twenty-five diverse bacterial isolates were separated from 25 waste water samples gathered from 15 diverse sites. The standard techniques were applied to obtain pure cultures [21, 22]. It was evident that only six bacterial strains could survive in the presence of 500 mg/L of 4-CP in media. Also, two bacterial strains namely, ChE_BE_1 and Che_BE_2, showed significant growth at 1,000 mg/L concentration of 4-CP.
The strain Che_BE_1 was chosen in the present study. The said bacterial stain was preserved by sub-culturing in petriplates enclosing inorganic medium supplemented with 1,000 mg/L of 4-CP and 1.5% agar. Stock culture was preserved at 4°C on a slant encompassing similar media. Che_BE_1 strain was initially identified as a gram-positive bacterium through biochemical characterization. To the best of our knowledge, Che_BE_1 is one of the potential isolate capable of metabolizing 1,000 mg/L of 4-CP in media and may serve as a potential candidate for the management of industrial wastewater contaminated with a high concentration of 4-CP.
3.2. 16S rDNA Gene Sequencing, Phylogenetic Analysis, and Thermodynamic Properties Analysis
16S rDNA technique was employed for genomic recognition of Che_BE_1 species, which was isolated from wastewater effluent. Genomic DNA of this strain was extracted, and it was amplified using PCR with a specific primer designed for 16S rRNA-specific. A single discrete PCR amplicon band of 1,500 bps was observed when resolved on 1.2% agarose gel (Fig. 1). The PCR amplicon was purified to remove contaminants. Forward and reverse DNA sequencing reactions of PCR amplicon were carried out with DF and DR primers using BDT v3.1 cycle sequencing kit on ABI 3730xl Genetic Analyzer. The consensus sequence of 1,470-bp16S rDNA was generated from forward and reverse sequence data using aligner software (Table S1). The consensus sequence of 16S rDNA gene was submitted to NCBI GenBank record (www.ncbi.nlm.nih.gov) with accession number of MF447840.1 [16].
The consensus sequence of 1,470-bp16S rDNA sequence was applied to perform BLAST amid NCBI GenBank record. Information concerning other adjacent homologs for microorganisms may be obtained in the alignment table (Table 1). The species ChE_BE_1 showed phylogenetically adjacent associated to Bacillus subtilis (B. subtilis), species ZAP018-1 (GenBank number: KU587801.1), B. subtilis, strain F3-3 (GenBank number: KT735214.1), B. subtilis, strain PCD3 (GenBank number: KY910140.1), B. subtilis, strain FL39 (GenBank number: KY818957.1), B. subtilis, strain IP18 (GenBank number: KY621529.1), B. subtilis, strain JL02 (GenBank number: KY400283.1), B. subtilis, strain JL12 (GenBank number: KY400272.1), Bacillus tequilensis, strain 7PJ-7 (GenBank number: KR708845.1), B. subtilis (GenBank number: KX058074.1), B. subtilis, strain D12-5 (GenBank number: KT955740.1), B. subtilis, strain VASB19/TS (GenBank number: KT427429.1), B. subtilis, strain 6R3-15 (GenBank number: LC155964.1), Bacterium, strain CDSHGTR2A-21 (GenBank number: KU743239.1) and Bacterium, strain CDSHGTGPM-16 (GenBank number: KU743237.1), and showed 98% sequence match. Hence, the isolated bacterial strain was recognized as B. subtilis. The highest possibility phylogenetic tree created employing MEGA 5 tool was exhibited in Fig. 2. The evolutionary background was measured applying the neighbor-joining method [23]. The bootstrap consensus tree anecdotal as replicates of 1,000 [24] was used to signify the evolutionary background of the taxa investigated [23]. The evolutionary span was computed applying the Jukes-Cantor technique [25] along with expressed in the units of the numeral of base changeover for each position. The rate difference amongst sites was computed using a gamma distribution (form factor = 1) [16]. The thermodynamic properties were obtained from the online tool (https://cail.cn/biotool/oligo/index.html). Results showed that B. subtilis MF447840.1 has 55% of GC content. Gibbs free energy (ΔG) and enthalpy (ΔH) along with entropy (ΔS) were found to be −2,464.4 kcal mol−1, −13,040.1 kcal mol−1 and 34,081.6 cal mol−1 K−1, respectively.
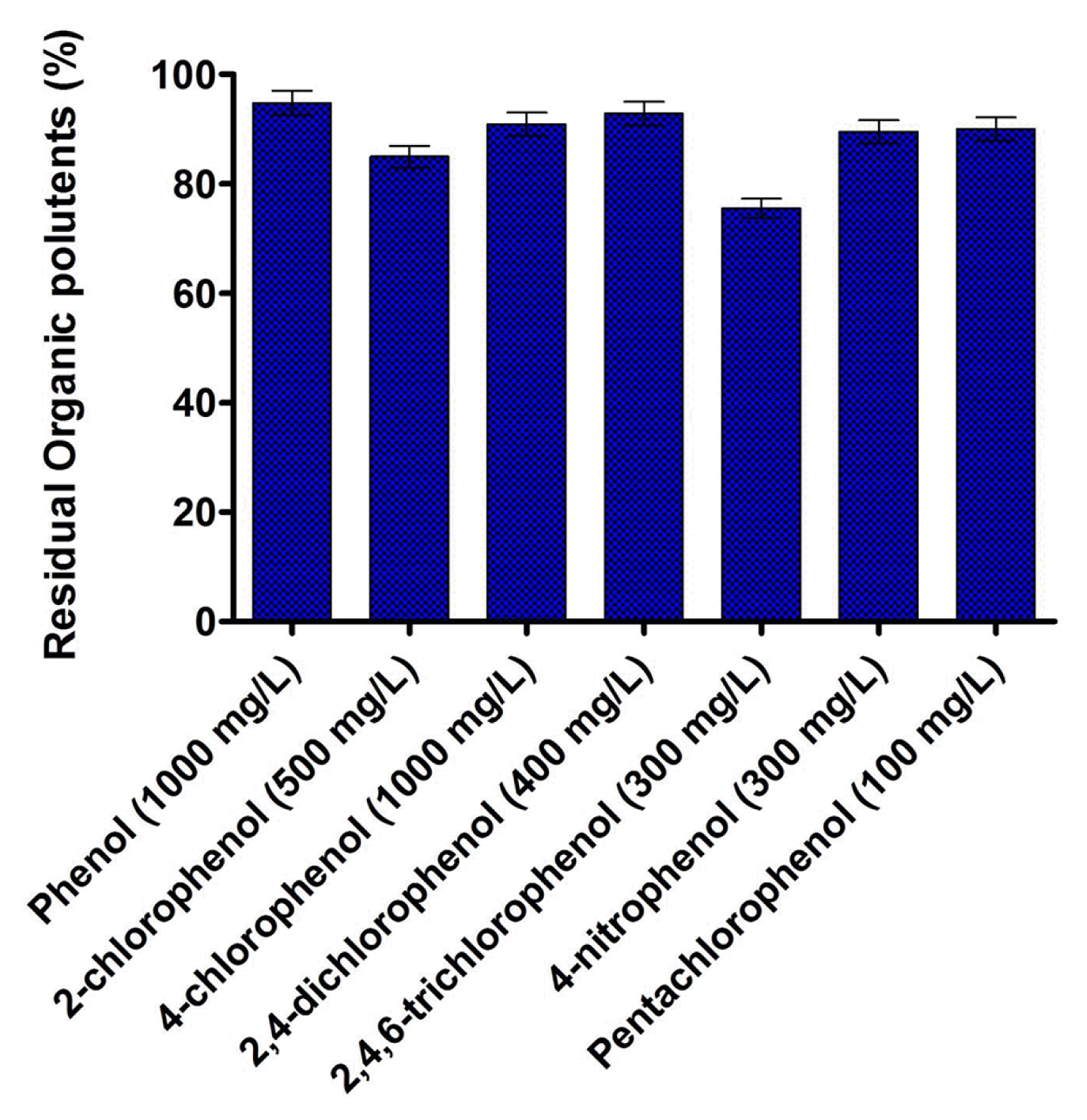
3.3. Metabolic Adaptability Analysis
The metabolic adaptability of B. subtilis MF447840.1 in presence various phenolic compounds is illustrated in Fig. 3. The results showed that the isolated strain could able to degrade 94.79% of phenol, 84.96% of 2-chlorophenol, 90.90% of 4-CP, 92.82% of 2,4-dichlorophenol, 75.53% of 2,4,6-trichlorophenol, 89.52% of 4-nitrophenol and 90.02% of pentachlorophenol after 48 h of incubation at 37°C. Fig. 3 illustrates that pentachlorophenol is found to be more toxic for isolated strain followed by 2,4,6-trichlorophenol, 4-nitrophenol, 2,4-dichlorophenol, 2-chlorophenol, 4-CP along with phenol. Since the degradation ability of various phenolic compounds and their utilization for growth is an intrinsic parameter of the microorganism, therefore, different degradation was observed after 48 h of incubation in inorganic media. It was archived that the species of Bacillus are well known for the degradation of phenolic compounds, however, in most cases, the degradation was found at lower concentration [26, 27]. Additionally, several other bacterial species, such as Pseudomonas sp. [28], Alcaligenes sp. [29, 30], Rhodococcus sp. [31], etc., can assimilate a wide variability of phenolic compounds, however diversity on the degradation of the various aromatic compound is limited. Therefore, researchers are looking for new effective microbial species which can able to degrade the wide variety of aromatic pollutants at higher concentration. As per our knowledge concern, B. subtilis MF447840.1 will be the first bacterial species which can be able to metabolize both 4-CP and phenol as well as five different aromatic compounds at higher concentration after 48 h of incubation. This metabolic adaptability creates B. subtilis MF447840.1 an effective microorganism intended for the bioremediation of an industrial effluent spoiled amid diverse sort of phenolic compounds.
3.4. Kinetics of Biodegradation of 4-CP
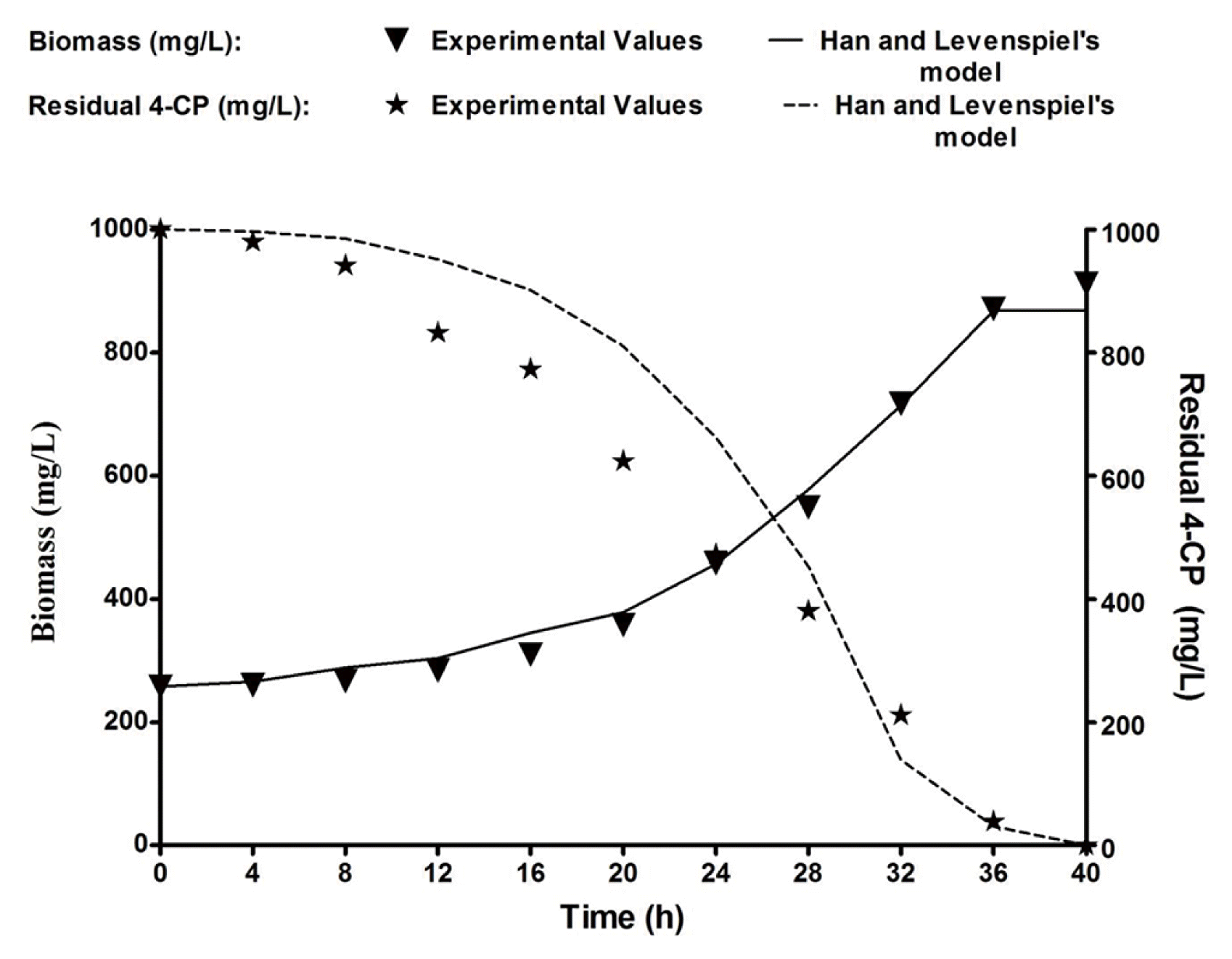
Biodegradation of 4-CP was conducted using B. subtilis MF447840.1 in batch cultivation with the optimized inorganic media supplemented with 1,000 mg/L of 4-CP. The experimental biomass data were plotted with time, and the same graph was used to plot the residual substrate (4-CP) (Fig. 4). The specific growth rate was calculated by applying Eq. (1), and the residual substrate was measured from Fig. 4. The specific growth rate (μ) of an isolated bacteria on diverse concentrations of 4-CP was calculated during the exponential phase using Eq. (1) [32].
Where, X is the cell concentration (g/L) at time t (h), and μ is the specific growth rate (h−1) [32]. The Monod model is usually applied to portray the association among the specific growth rate (μ) and the concentration of the restraining substrate using Eq. (2). Generally, Monod or Haldane models have been used to characterize the biodegradation kinetics of several phenolic compounds. However, in some cases, these equations are not adequate to represent the biodegradation process under transient conditions [33].
Where, μmax is the highest specific growth rate (h−1), KS is the saturation constant (g/L), and S is the substrate concentration (g/L). In this study, GraphPad Prism 5 was applied to resolve the nonlinear equation applying non-linear regression analysis. Numerous kinetic factors, namely, μ, μmax, and KS were computed by fitting the experimental data. A plot of μ versus substrate concentration designated the substrate inhibition happening in the concentration range of the substrate investigated (Fig. 5). Consequently, an effort was made to fit the experimental data to the obtainable kinetic models on substrate inhibition. Various substrate inhibition kinetic models were investigated and compared in the present work (Table 2) [34–37]. The specific growth rate (μ) was plotted with substrate concentration in Fig. 5 to determine various kinetic parameters. By trial and error method, the experimental data were shown to fit reasonably well in Han and Levenspiel’s model [36] for cell growth and 4-CP utilization. The model suggested that the 4-CP degradation rate showed an impediment role, which had an exponential form along with combined the substrate concentration corresponding to the disparity position on growth [36, 38].
In Han and Levenspiel’s model described in Eq. (3), μ and μmax are the specific growth rate (h−1) and highest specific growth rate (h−1), respectively. Sm is the decisive inhibitor concentration (mg/L) above this concentration the bacterial growth stops. The m and n are the experimental constants. The values of kinetic parameters such as μmax and KS were obtained to be similar to 0.11 h−1 and 39.88 mg/L, respectively. Critical inhibitor concentration for B. subtilis MF447840.1 was found to be 1 g/L. The values of m and n were 1.57 and 1.046, respectively, while the correlation coefficient (R2) was 0.9993. The higher correlation coefficient value of Han and Levenspiel’s model indicates the suitability of the model as compared to other tested substrate inhibition models in the present study. Yx/s value is calculated by plotting ΔX versus ΔS at various time intervals. The slope of this straight line equation indicates the magnitude of Yx/s. From the experimental value, Yx/s was measured and reported as 0.53 g of biomass/g of 4-CP. As 4-CP is environmentally toxic, the value of Yx/s obtained herein this study was minuscule. Generally, the Yx/s value for bacterial cultures is within the array of 0.6 to 0.72 g/g [39]. However, the yield value was 0.65 g/g for Pseudomonas putida in the existence of phenol as the singular carbon basis [40]. As Yx/s is an intrinsic parameter of microorganisms, the values may be different for various microorganisms. Also, the yield also depends on types and concentrations of substrates and environmental parameters [41]. It has reported that 4-CP is degraded by an extracellular enzyme secreted by the microorganism. Although the experimental data were collected throughout 40 h to evaluate the degradation efficiency of B. subtilis, however, the log phase data were only taken into account for the calculation of Yx/s. 4-CP is considered as a limited substrate required for growth. Furthermore, maintenance was not considered during the calculation of Yx/s, as it is considered to be low during the log phase [32]. The simulated data of biomass were calculated using Eq. (3) and used to achieve the predicted residual amount of 4-CP using Eq. (4).
Where, YX/S is yield coefficient (g of biomass produced/g of 4-CP), and –dS/dt is the degradation rate (gL−1h−1). Both experimental and predicted values of biomass and residual 4-CP were illustrated in Fig. 4. Fig. 4 shows that the experimental data are reasonably fit with predicted data, indicating that the Han and Levenspiel’s model is appropriate for the present case.
Where, q and μ are the specific degradation rate of 4-CP (h−1) and specific growth rate (h−1), respectively. The simulated data of μ was calculated using Eq. (3) and used to achieve the predicted q using Eq. (5). Both investigational along with envisaging data of μ and q were illustrated in Fig. 5(a) and (b). Fig. 5 shows that the predicted data using Han and Levenspiel’s model are reasonably fit with experimental data, indicating that the said model is suitable for the present study. Table 3 indicates the degradation efficiency of 4-CP using various microorganisms. As per our knowledge, there is no microorganism discovered till date which can degrade 4-CP at 1,000 mg/L concentration. Jiang et al. [42] investigated the biodegradation of 4-CP using the mutant strain Candida tropicalis (C. tropicalis) CTM 2 under batch condition. It is evident that 400 mg/L of 4-CP was degraded by the mutated strain within 59.5 h. Basak et al. [43] used C. tropicalis PHB5 to evaluate the biodegradation of 4-CP and found that 949.7 mg/L of 4-CP was entirely degraded after 60 h in batch culture. In the present study, 1,000 mg/L of 4-CP was entirely degraded by B. subtilis MF447840.1 within 40 h. Therefore, B. subtilis MF447840.1 may be considered as a potential candidate for the degradation of 4-CP.
4. Conclusions
Twenty-five different bacteria were isolated from twenty-five waste water samples which were collected from fifteen different places. The phylogenetic study was performed to identify the most potent 4-CP-degrading strain. Genomic identification showed that the isolated strain was phylogenetically associated amid B. subtilis having GenBank accession number MF447840.1. The in-depth kinetic investigation was performed to assess the effect of various concentrations of 4-CP on the growth of B. subtilis. The intrinsic kinetic parameters were computed using a non-linear regression analysis with best fit unstructured Han and Levenspiel’s model. Experimental data on substrate concentration dependency of the specific growth rate in this study fit well with the simulated data find by using the best fit unstructured Levenspiel’s equation. B. subtilis MF447840.1 is the first bacterial species which metabolized both 4-CP and phenol at the concentration of 1,000 mg/L as well as five different aromatic compounds. This metabolic adaptability creates B. subtilis MF447840.1 a potent microorganism for the biodegradation of industrial effluents contaminated with various types of phenolic substances. Thus, the present study may expose new heights in research in the areas of biological treatment of industrial effluents containing various phenolic compounds. The microorganism isolated and used in this study for 4-CP biodegradation is a competent one and can further be exploited in industrial scale applications.