1. Introduction
Harmful algal blooms (HABs) are the rapid growth of microscopic algae that can cause harm to animals, people, and the local environment. Thus, determining the physiological status of algal growth is important for effective water quality and algal bloom management in freshwater environments [1]. Particularly, Microcystis sp. is one of the most common harmful cyanobacteria found in freshwater [2–6]; they produce toxic substances, namely microcystins, which are known to cause serious damage to the human liver [7–9]. Proper management of Microcystis sp. is important for securing safe water for the public [7, 10–12]. Biomass, cell density, and chlorophyll-a concentrations are often measured to estimate the status of algal growth [1, 13–17]. Counting algal cells is also important for determining the status of eutrophication in water bodies worldwide, as cell number is often included in guidelines for algal regulation, such as the World Health Organization guideline [18–20]. For example, in South Korea, Microcystis sp. is regulated based on the total number of cells within a colony of Microcystis sp. (cells/mL). Microscopic investigation is widely used for counting algal cells; however, it is a time-consuming, subjective, and labor-intensive method. Given that HABs are an increasing global issue, it is urgent to develop a simple, rapid, and accurate analytical method for enumerating the individual cells in an algal colony.
The FlowCAM (Flow Cytometer and Microscope, Fluid Imaging Technologies, Yarmouth, ME, USA) is a potential alternative to the conventional cell counting method [21] that has been widely used for the identification, classification, and biomass measurement of algae in water bodies by providing unbiased results within a relatively short time (e.g., within 5 min) [22–24]. Rowe et al. captured images of individual colonies of Microcystis sp. in western Lake Erie, United States using a FlowCAM, and estimated their equivalent spherical diameters using image analysis [4]. The FlowCAM is a particle image analyzer that was originally developed for the analysis of various particulate matters in water, including zooplankton and phytoplankton [25–28]. It identifies algae based on shape and morphological parameters of algal images, such as length, width, area, area-based diameter, and equivalent spherical diameter, which are obtained from a microscope and a digital camera equipped in the FlowCAM imaging platform [23, 25]. By processing FlowCAM images, a photo image library for algal samples is generated by users, and the characterized shapes and morphological parameters of algal images are used for the proper identification and classification of algal species in unknown water samples [24]. Thus, the acquisition of discriminable image pools of target algal species is essential for accurate measurement of algal cell counts.
Although the FlowCAM provides valuable information on algal communities, there are still limitations in counting individual cells within a Microcystis sp. colony, which is essential for the management of drinking water resources. The FlowCAM analyzes two-dimensional (2D) surface areas. If algae are not distributed in the form of single cells (e.g., algal aggregates or clumps), then this 2D cell counting method will be biased. This may result in underestimation of the total number of algal cells. To overcome this, Wang et al. [14] employed sonication or alkaline hydrolysis to disintegrate the algal colonies, and measured unicellular Microcystis using a FlowCAM. However, there is still a lack of image processing techniques, which are required to improve the accuracy of algal cell measurements using a FlowCAM.
To the best of our knowledge, there is no standard procedure for counting the individual cells in a 3D Microcystis colony using a FlowCAM. In this study, we developed a novel method for automated quantification of Microcystis cells in fresh water using a FlowCAM. The purpose of this study was to verify the applicability of the method in practical cell counting of Microcystis colonies for water quality management rather than to provide specific parameters of the developed method. First, Microcystis colonies within a water sample were identified using a FlowCAM with a local FlowCAM image library of Microcystis colonies collected in this study from fresh water of rivers and reservoirs in South Korea. Second, the total sum of the areas of the Microcystis colonies within the water sample was obtained from the FlowCAM, and then a model algorithm (i.e., the relationship between the number of cells vs. the 2D surface area of the algal colonies) developed in this study was applied to estimate the total number of individual cells of Microcystis sp. within the water sample. The accuracy of the model algorithm in counting the total number of individual cells in Microcystis colonies within the water sample was then evaluated by comparing the result from the FlowCAM with the result from a conventional microscopic method for algal cell counting.
2. Materials and Methods
2.1. Development of a FlowCAM Image Library for Microcystis sp
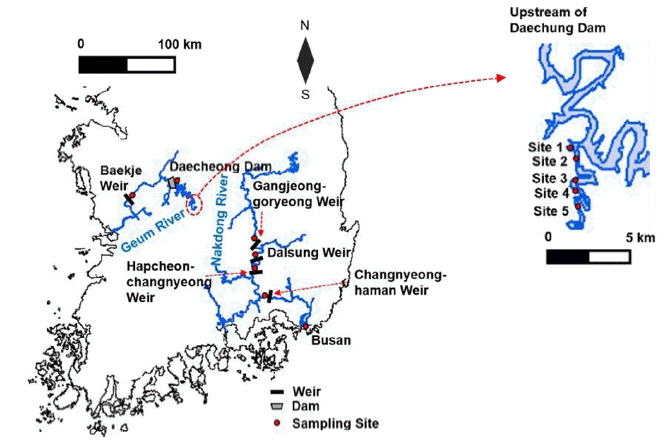
Currently, there are no sufficient FlowCAM libraries available for the identification of Microcystis sp. found in major rivers in South Korea; thus, in this study, a specific image library of Microcystis sp. from Korean rivers was developed. A FlowCAM (8100-C, Fluid Imaging Technologies Inc., Scarborough, ME, USA) was used to generate the image library of Microcystis sp. in water samples collected from two of four major rivers, namely the Nakdong River and the Geum River, in South Korea between June 27 and August 16, 2017 (Fig. 1 and Table 1(a)), where algal blooms have been an important issue in water quality management [29]. The water samples were collected from the surface of the two rivers.
The Nakdong River is the longest river in South Korea with a flow path length of 510 km and a watershed area of about 23,000 km2. There are eight weirs in the middle of the river, which were built in 2011. The Geum River is the third longest river in Korea with a flow path of length of 400 km and a watershed area of about 9,885 km2. The Daecheong Dam is located about 135 km upstream of the mouth of the Geum River, and has a storage capacity of 1.5 billion tons and a watershed area of 4,100 km2. It was built in 1980 to provide drinking water and to manage floods during the summer season.
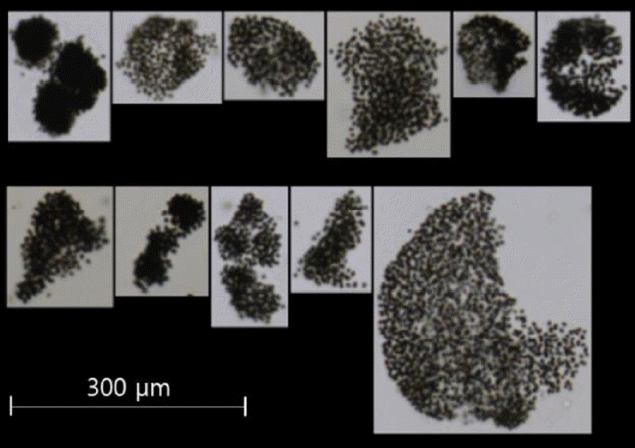
A total of 2,520 photo images of Microcystis colonies were obtained using a FlowCAM from the samples that were randomly collected from four sites in the Nakdong River, namely the Dalsung Weir (July 25, 2017), Hapcheon-changnyeong Weir (June 27, 2017), Changnyeong-haman Weir (June 27, 2017), Busan (July 19, 2017), and one site at about 35 km upstream of the Daecheong Dam (August 16, 2017) in the Geum River (Fig. 2). The FlowCAM identifies algal images when a water sample flows through a filter tube inside the device where the diameter of the filter tube is coupled with a specific magnification (e.g., 300 μm tube filter with ×400 magnification or 600 μm tube filter with ×200 magnification) for cell counting. In this study, 300 μm tube filter connected with a microscope (×400) was used as it typically covers most dominant sizes of colonies found in water. The colonies larger than 300 μm in diameter were filtered to prevent clogging in the FlowCAM filter tube, and thus were excluded from the library. Approximately 60% of the photo images (1,500) were taken from the samples collected in the Nakdong River, and the rest were from the samples collected in the Daecheong Dam. The 2,520 photo images were utilized for the development of a library of FlowCAM images that was used for the identification and classification of Microcystis colonies in unknown water samples.
2.2. Colony Surface Areas and Number of Cells in a Colony
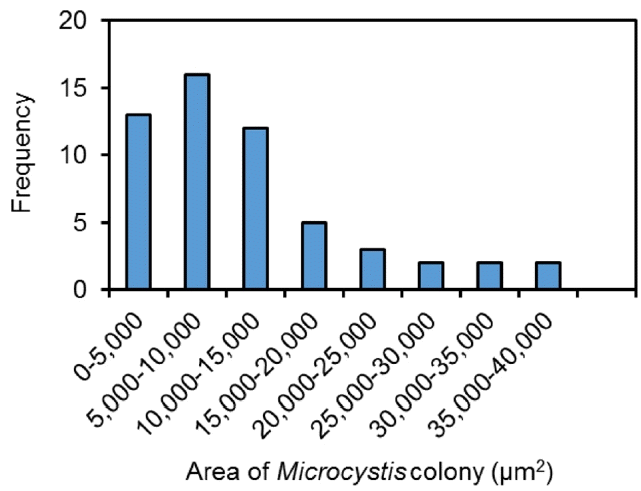
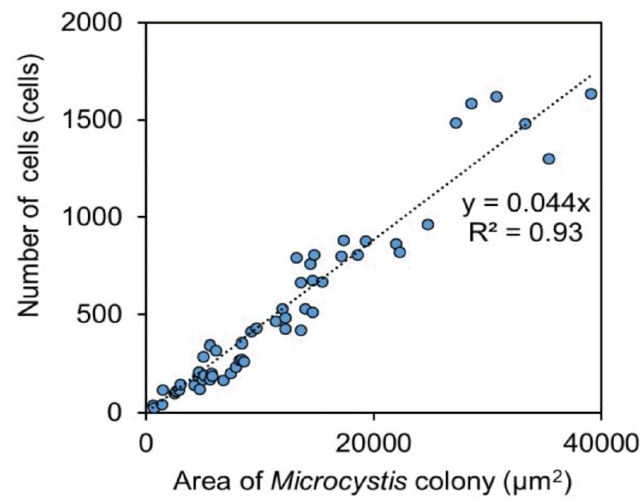
Water samples collected from the Daecheong Dam (Site 4) in the Geum River between August 16 and 17, 2017 were used to develop a model algorithm for the cell count of Microcystis colonies (Fig. 1). A total of 55 Microcystis colonies were analyzed to determine the relationship between the 2D surface area of a colony and the number of cells in each colony. For visual identification of cell numbers using a conventional microscopic method, individual Microcystis sp. cells were counted under a microscope (×400, Nikon DS-Ri2, Tokyo, Japan). It should be noted that this method counted only individual cells, which were identified under a microscope with 2D images, and was expected to underestimate the actual number of cells. The area of a single Microcystis colony ranged from 579 μm2 to 39,029 μm2 (Fig. 3). Then, a model algorithm was developed from the relationship between the number of cells and the area of the colony.
2.3. Determination of Relationship between FlowCAM Images and the Cell Counting Model Algorithm
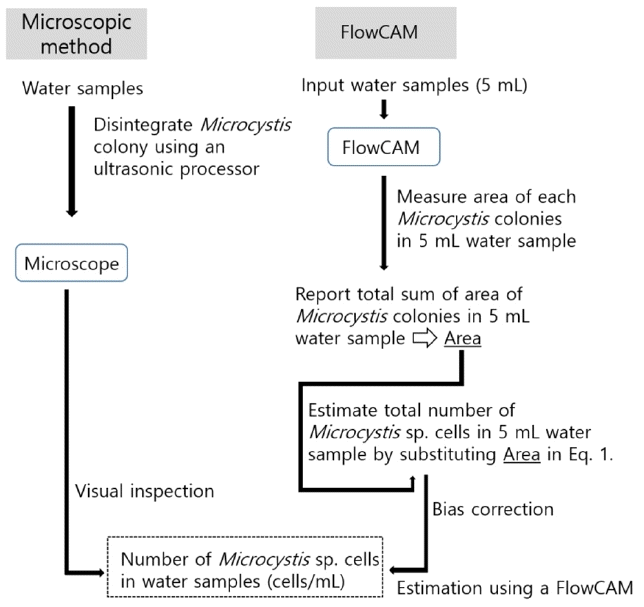
For cell number counting by FlowCAM analysis, the FlowCAM was first used for the identification of Microcystis colonies in a water sample (e.g., 5 mL). It then calculated the surface area of the 2D Microcystis colony and determined the cell numbers using the relationship between the number of cells in the Microcystis colony and the surface area of the Microcystis colony, which was developed by counting the cells in each colony in the 2D image, as explained in Section 2.2.
To evaluate the developed model algorithm for Microcystis sp. cell counting, unknown water samples randomly collected from the two rivers (Fig. 1 and Table 1(b)) were analyzed by comparing cell numbers between a conventional microscopic method and the FlowCAM method (Fig. 4). A total of 26 samples collected from the Daecheong Dam in the Geum River (August 16, 17, and 31, 2017), one sample from the Baekje Weir in the Geum River (September 7, 2017), and one sample from the Gangjeong-goryeong Weir in the Nakdong River (August 8, 2017) were separately used to evaluate the uncertainty of the developed model algorithm. For weirs, water samples were collected about 500 m upstream of each weir body. Water samples at the Daecheong Dam were randomly collected from five different sites between 30 and 36 km upstream of the dam body. Five milliliters of each sample were analyzed using the FlowCAM, and the number of cells (cells/mL) was compared between the FlowCAM method and visual inspection with a conventional microscope to determine the accuracy of the model algorithm developed in this study.
The conventional microscopic method measured only the cells that were identifiable in 2D. Thus, individual cell counting of a Microcystis colony was almost impossible without breaking the colony because the colony was a 3D shape. For visual identification in this step, individual Microcystis cells were counted using a conventional microscopic method using a Sedgewick-Rafter counting chamber after disintegration of the Microcystis colony by sonicating 1 mL of the water sample for 10–20 min at 28 kHz and 80 W using an ultrasound device (Ultrasonic Processor CS-1025, Chosun 21, Seoul, South Korea) with the addition of 100 μL of 1 M NaOH [14, 30, 31]. The sonification time was determined based on preliminary tests by monitoring Microcystis colony disintegration under the conventional microscope after sonication (28 kHz and 80 W) with different durations.
3. Results
3.1. Relationship between Number of Cells in a Colony and Colony Areas
The relationship between the number of cells in a Microcystis colony and the surface area of the colony is shown in Fig. 5. A linear regression model (Eq. (1)) showed a relatively strong relationship between cell numbers and surface area measured by visual inspection using microscopy, with a coefficient of determination (R2) of 0.93.
where surface area is the surface area of a Microcystis colony in a water sample (μm2). As the number of cells should be zero when the area of a colony is zero, the intercept of the model equation was determined as zero.
3.2. Bias Correction between the Two Methods and Development of a Model Algorithm
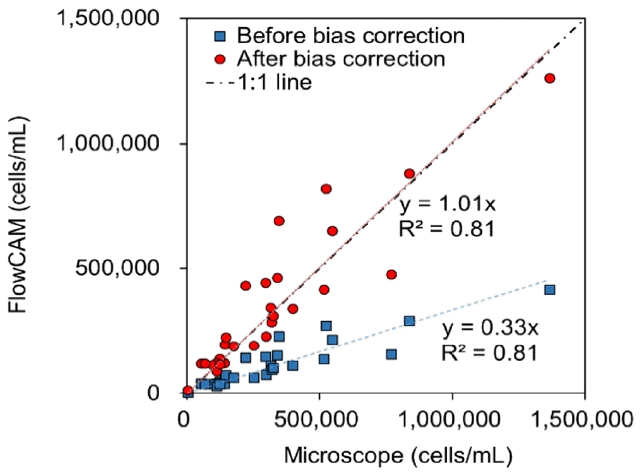
The number of cells determined by the FlowCAM underestimated the actual cell numbers in a 3D colony. For a simple, fast, and accurate analysis using a FlowCAM, the difference between the cell numbers from the FlowCAM analysis and the actual cell numbers should be determined. To count the actual cell numbers using the conventional microscopic method, sonication was applied to break algal clumps. The number of Microcystis sp. cells was compared between the FlowCAM and the conventional micro scopic method after sonication (Fig. 6). It was found that the number of cells determined by the FlowCAM (plotted as blue squares in Fig. 6) underestimated the actual cell numbers after sonication, as the number of cells in a single Microcystis colony estimated from Eq. (1) represented the number of cells within the Microcystis colonies limited to 2D images. The slope of the relationship between measured cell numbers from the FlowCAM and microscope was 0.33 with an R2 of 0.81.
To compensate for the loss of the actual cell numbers in cell counting, correction of this bias was needed. The correction factor was determined from the relationship of a 1 to 1 line and the slope of the relationship between the cell numbers measured by the FlowCAM and cell numbers measured by the microscopic inspection; the correction factor was determined as 1/0.33 = 3.03. Thus, the correction factor of 3.03 was multiplied by the cell numbers originally estimated by the FlowCAM to correct the actual cell numbers. The corrected cell numbers measured by the FlowCAM are plotted as red circles in Fig. 6, where the slope of the relationship of measured cell numbers from the FlowCAM and microscope was 1.01.
3.3. Model Validation with Statistical Analysis
The accuracy of the developed model algorithm was evaluated using three statistical methods, namely root mean square error-observation standard deviation ratio (RSR), Nash-Sutcliffe efficiency (NSE), and R2, using Eq. (2)–(4), which are commonly used for the evaluation of model performance [32, 33].
where n is the number of observations, Qi is the ith observed value, Ō is the mean of Oi, Mi is the ith model-estimation value, and M̄ is the mean of Mi.
The performance of the presented model algorithm (i.e., estimation of Microcystis sp. cell numbers in the water sample combining Eq. (1) and bias correction, as shown in Fig. 6) was analyzed based on cross-validation of observed and predicted cell numbers using RSR, NSE, and R2. For each iteration, 27 out of 28 samples were used for model calibration, and the remaining sample was used for validating the model algorithm; this step was iterated 28 times to measure residuals between observed data and model predictions for each of the samples. The results of the model performance are presented in Table 2. The average values of RSR, NSE, and R2 of the FlowCAM model were 0.44, 0.80, and 0.80, respectively. Better model performances are achieved if the values of RSR are closer to zero and NSE and R2 values are close to 1; the model estimation can be considered satisfactory when RSR < 0.7 and NSE ≥ 0.65 [32, 33]. Therefore, it was concluded that the developed model algorithm for cell number counting using the FlowCAM in this study was well calibrated, and a strong correlation was achieved between the observed and predicted cell numbers for the 28 samples.
4. Discussion
The manual measurement of Microcystis sp. using a microscope is a time-consuming process that requires highly trained expertise to avoid possible bias during visual inspection [24]. However, for effective water quality management, especially during algal bloom events such as those during the summer season, fast sampling and rapid analysis are required to make a quick, responsive decision to control algal bloom events properly. It is known that FlowCAMs can provide fast and unbiased results, which is valuable for water resource management regarding HABs in reservoirs and rivers [24, 25]. However, the quantification of algal blooms using FlowCAMs is still in the early stages of investigation, and the correction for accurate cell counting is not well explored. One challenge is counting cells in a 3D colony using a 2D analytical method. This study is the first to provide a systematic procedure and to develop a model algorithm for cell counts of Microcystis colonies considering the 3D form of the colony. The cross-validation of the developed model algorithm with unknown samples using the FlowCAM showed satisfactory performance for the estimation of Microcystis sp. cell numbers, which is considered adequate for practical application for rapid detection and analysis of Microcystis blooms in reservoirs and rivers with the following considerations.
First, the purpose of this study was to provide a new approach to facilitate cell counting of Microcystis sp. using a FlowCAM with a simple, but accurate analytical model and procedure, rather than to suggest specific values in the algorithm. For the water samples tested in this study, the bias correction factor (3.03) was determined based on the relationship of a 1 to 1 line and the slope of the relationship between cell numbers obtained by the FlowCAM and by microscopic observation. As samples randomly collected from various watersheds and sites were utilized for modeling, the model algorithm in this study could be considered generally applicable for HABs containing Microcystis sp. as predominant algal species. However, algal species and aggregation may be site-specific and thus FlowCAM library and model algorithm such as a slope in Eq. (1) or a bias correction factor (3.03 in this study) need to be developed initially for the FlowCAM use and the results may be different depending on sites. Using the proposed model algorithm and analytical procedures from this study, one can easily develop their own FlowCAM library and algorithm and utilize the FlowCAM for accurate cell counting for different sites and watersheds. Geomorphological comparison of a 3D ball and a 2D circle and further analytical analysis of the size of individual Microcystis sp. cells may also provide an alternative approach to improve the model algorithm.
Second, in this study, a filter tube with a diameter of 300 μm coupled with a microscope of ×200 magnification was used for algal cell imaging and counting. Although some of Microcystis colonies larger than 300 μm may enter the filter tube by flow, the detection of Microcystis colonies was typically limited to colonies with a diameter less than 300 μm. Larger colonies can be detected using a next available filter tube larger than 300 μm which is a 600 μm filter tube with a microscope of ×200 magnification. However, the use of different filter tubes with different magnifications for sample analysis can make the system more complicated with increased uncertainties, requiring further study. In practice, 300 μm tube filter with a microscope of ×400 magnification is typically used as it covers most dominant sizes of colonies found in water.
Lastly, the preciseness of the FlowCAM image library for the identification of Microcystis sp. was not evaluated in this study. The accuracy of the FlowCAM in the classification of Microcystis colonies itself is dependent on the appropriateness of the Microcystis sp. image library, which is individually developed by users. The development of an improved library could improve the applicability of the algorithm developed in this study to count the number of cells in Microcystis colonies.
5. Conclusions
In this study, a novel method for cell counting in Microcystis colonies was developed from water samples collected from two major watersheds in South Korea. The number of cells in Microcystis colonies obtained from the model algorithm was evaluated by cross-validation with three statistical methods, where the average values of RSR, NSE, and R2 of the FlowCAM model were 0.44, 0.80, and 0.80, respectively. This study demonstrated the practical applicability of the FlowCAM for rapid and unbiased analysis of algal blooms, which is important for the proper management of water quality in freshwater systems and to provide safe water to the public. Further improvement of the precision of the FlowCAM library and the model algorithm for cell counting in algal colonies, as well as the extension of the library to other algae species, were left for future research.